
Very excited to share our latest work in Science on metagenomic editing (MetaEdit) of the gut microbiome in vivo & directly modifying unculturable immune-modulatory SFB bug in the small intestine. 🦠🧬🛠️
www.science.org/doi/10.1126/...
@lauralvllo.bsky.social
PhD student at Evodynamics lab (Hospital Ramón y Cajal). New strategies against antibiotic resistant bacteria and AMR evolution in bacteria.

Very excited to share our latest work in Science on metagenomic editing (MetaEdit) of the gut microbiome in vivo & directly modifying unculturable immune-modulatory SFB bug in the small intestine. 🦠🧬🛠️
www.science.org/doi/10.1126/...

Dissecting the fitness effects of a carbapenem resistance plasmid in clinical Enterobacterales using plasmid-wide CRISPRi screens
@aliciapcv.bsky.social @jorgesastred.bsky.social @sanmillan.bsky.social @cnb-csic.bsky.social
#bacteria #plasmids

Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n

New preprint from the group: @lucydillon.bsky.social analysis of 16,000+ genomes finds Bacteria cannot combine certain resistance genes as they are mutually exclusive, forcing them down incompatible evolutionary paths.
biorxiv.org/content/10.1101/2025.08.20.671315v1
@jomcinerney.bsky.social

New paper alert! 🚨
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇

Our paper “Genetic determinants of pOXA-48 plasmid maintenance and propagation in Escherichia coli” is now online in @natcomms.nature.com ✨
👉 www.nature.com/articles/s41...
@cnrs-rhoneauvergne.bsky.social @mmsb-lyon.bsky.social

First of all, we recommend reading a complementary study led by Yannick Baffert and @sbigot.bsky.social, in which they use Tn-seq to study the genetic determinants of pOXA-48 in plasmid maintenance and conjugation. Really cool piece of work that reaches similar conclusions to ours!
2/14
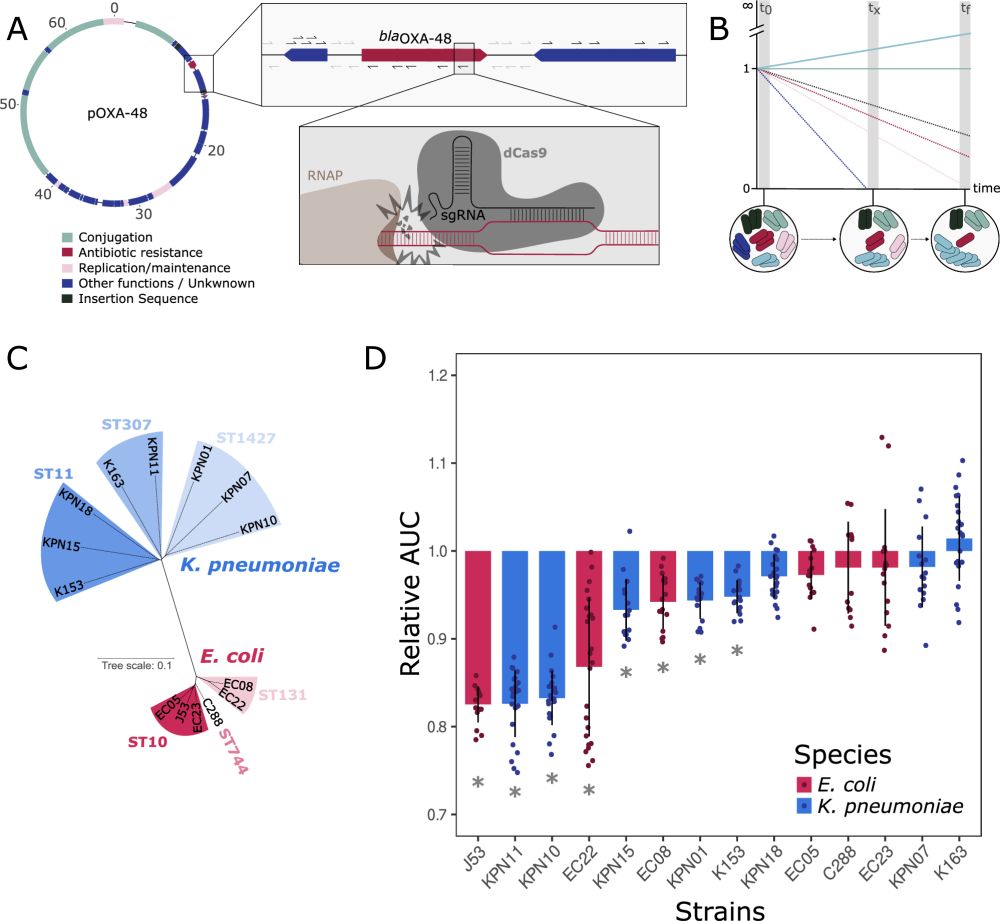
This work is finally published! 🥳🧬
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
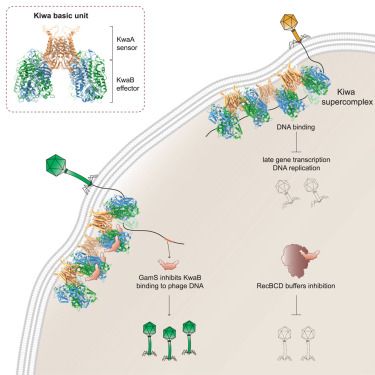
Thrilled to see our Kiwa story out today! A membrane-associated supercomplex that senses infection and blocks replication and transcription.
www.cell.com/cell/fulltex...
Huge congratulations to Yi and Zhiying for bringing it home, to Thomas for starting us off, and to all the collaborators.
#phage #phagesky
Phage-based delivery of CRISPR-associated transposases for targeted bacterial editing | PNAS www.pnas.org/doi/10.1073/...

#phagesky
www.cell.com/cell-host-mi...
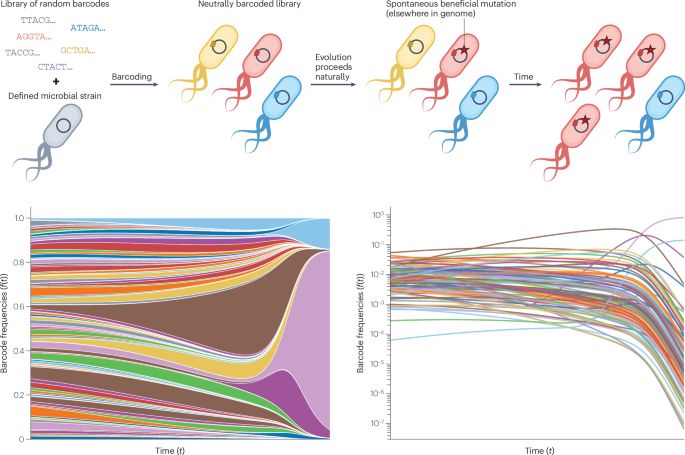
ICYMI: New online! Experimental evolution in an era of molecular manipulation
28.07.2025 14:46 — 👍 13 🔁 9 💬 0 📌 0Excited 🥳 to share our latest work on gut phages!
Big thanks to @epcrocha.bsky.social, Erick D, Camille d'H, @fplazaonate.bsky.social, Quentin LB, and all others involved for support and contributions! 🙌
Out in Cell Reports @cp-cellreports.bsky.social
doi.org/10.1016/j.ce...
Here's what we found 🤓
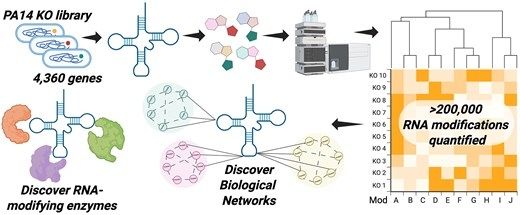
tRNA modification profiling reveals epitranscriptome regulatory networks in Pseudomonas aeruginosa 🦠👏High-throughput tRNA epitranscriptome profiling using mass spec on P. aeruginosa transposon insertion library and links with metabolic networks #microsky #rnasky doi.org/10.1093/nar/...
28.07.2025 20:28 — 👍 11 🔁 6 💬 0 📌 0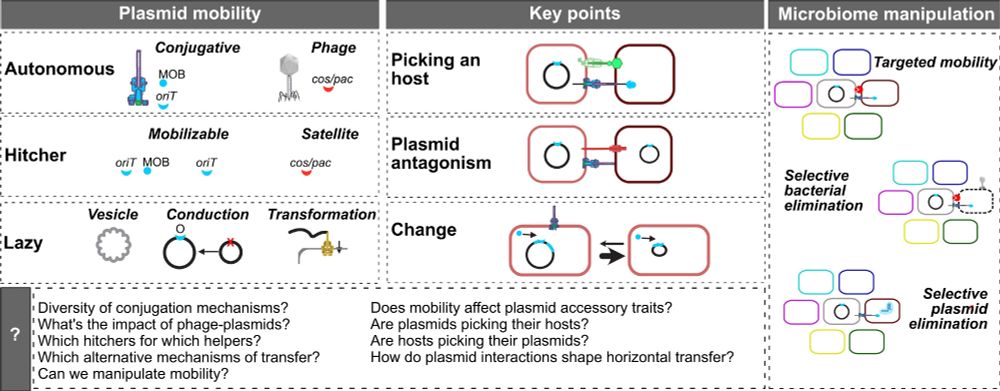
graphical abstract of the article the extended mobility of plasmids
Here's our new broad review on the extended mobility of plasmids, about all mechanisms driving and limiting their transfer. From conjugation to conduction, phage-plasmids to hitchers, molecular to evolutionary dynamics, ecology to biotech. The state of affairs. 1/9 academic.oup.com/nar/article/...
23.07.2025 07:35 — 👍 184 🔁 93 💬 4 📌 9

Had an amazing time at #FEMS2025 last week, where I presented our latest findings on collateral sensitivity mediated by beta-lactamases ☯️ Big thanks to everyone who came by! So proud of my colleagues for their brilliant talks and posters ✨
23.07.2025 06:43 — 👍 4 🔁 1 💬 0 📌 0The perfect summer read is here! 👇🏽
23.07.2025 06:17 — 👍 1 🔁 0 💬 0 📌 0A must read paper! 👇
02.07.2025 12:18 — 👍 4 🔁 0 💬 0 📌 0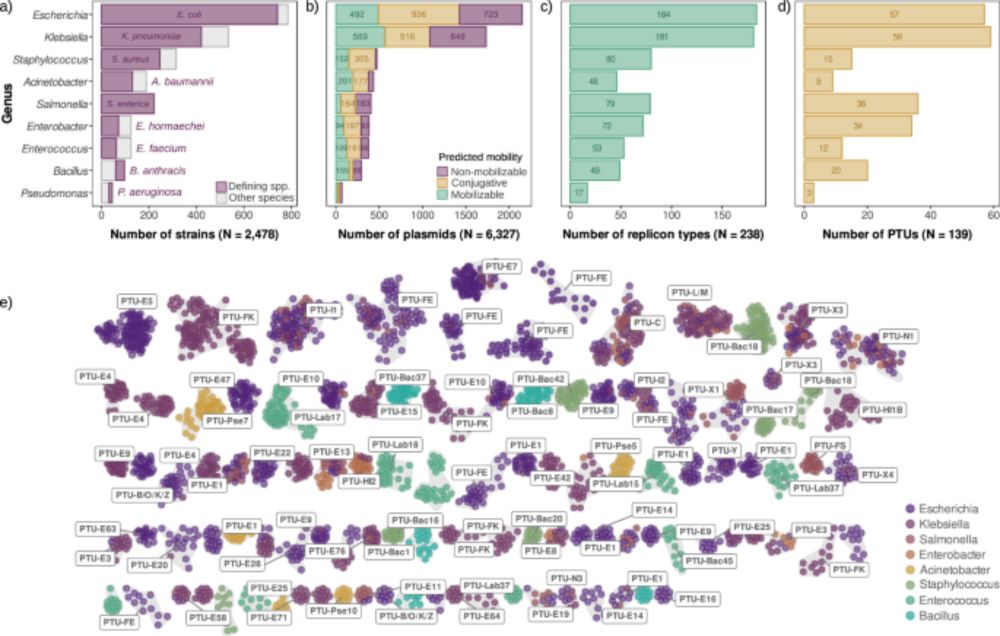
🚨🚨New paper out in @natcomms.nature.com!!
Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
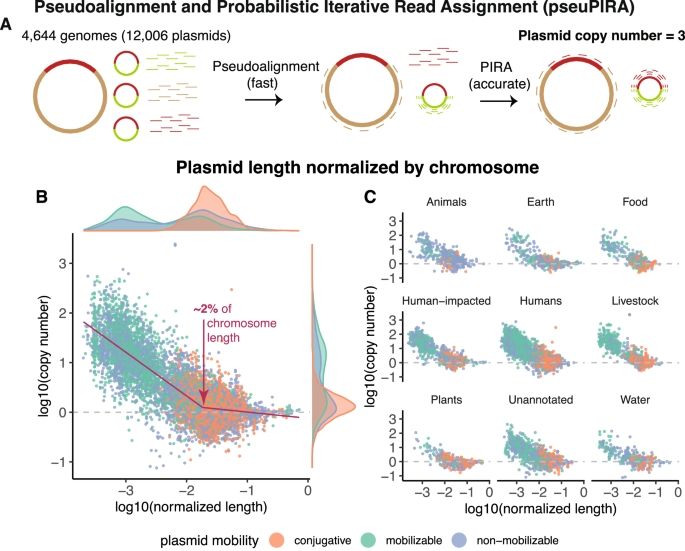
Scaling laws of bacterial and archaeal plasmids www.nature.com/articles/s41...
02.07.2025 11:35 — 👍 6 🔁 5 💬 0 📌 0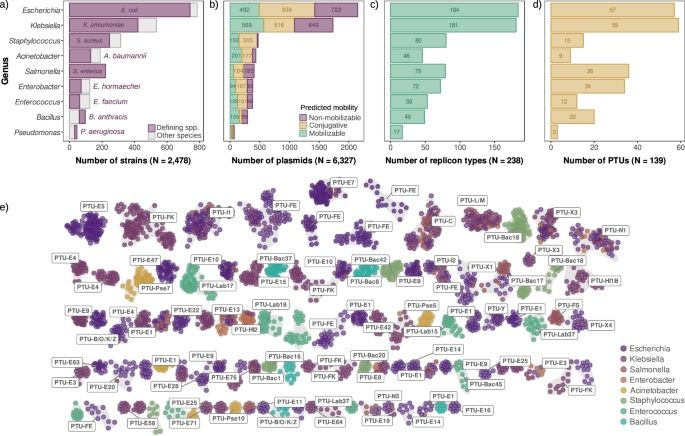
Universal rules govern plasmid copy number www.nature.com/articles/s41...
02.07.2025 11:37 — 👍 3 🔁 3 💬 0 📌 0
#NatMicroPicks
E. coli making paracetamol from plastic waste! 🦠💊
A new biocompatible chemistry for plastic waste upcycling using microbes.
#MicroSky #SynBio 🔧🧬
www.nature.com/articles/s41...

What determines who a phage can infect?
We tackled this question for temperate phages of Klebsiella — a bacterial pathogen — using a genome-wide association study (GWAS) and a massive protein testing effort.
👇 A thread!
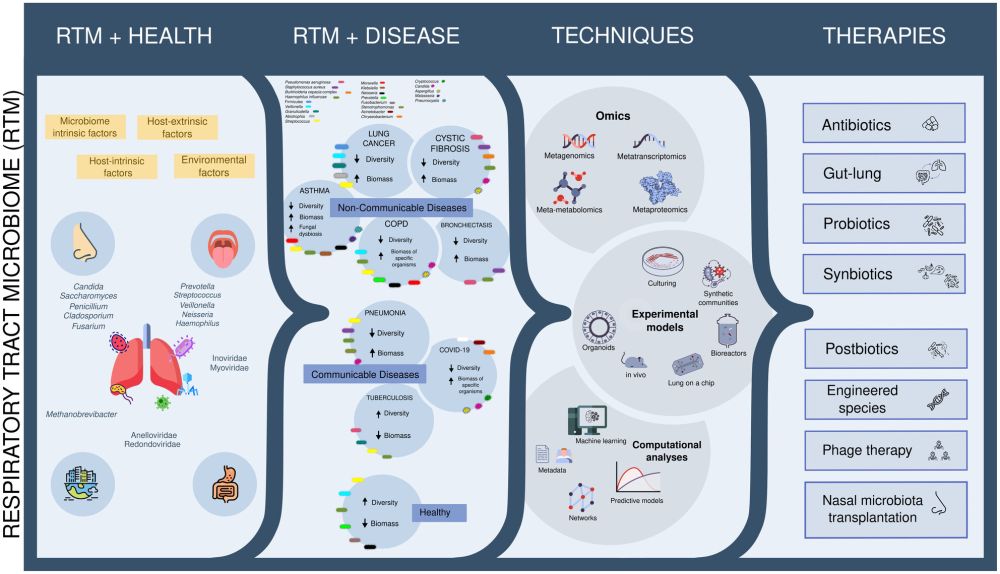
New mini-review on the respiratory microbiome and its contribution to human health from biotechnology 🫁🦠
@tmcoque.bsky.social @microbiotech.bsky.social
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...

New antibiotic class discovered by researchers at @mcmasteruniversity.bsky.social & @illinoispress.bsky.social
"A broad-spectrum lasso peptide antibiotic targeting the bacterial ribosome"
in @nature.com
Hope it can make it to market to help combat #AMR infections
www.nature.com/articles/s41...
The Eligo team did it again! We just shared on biorxiv the work we did towards the development of a CRISPR-Cas therapy targeting Shiga Toxin producing E. coli (STEC). www.biorxiv.org/content/10.1...
04.03.2025 12:31 — 👍 44 🔁 27 💬 1 📌 3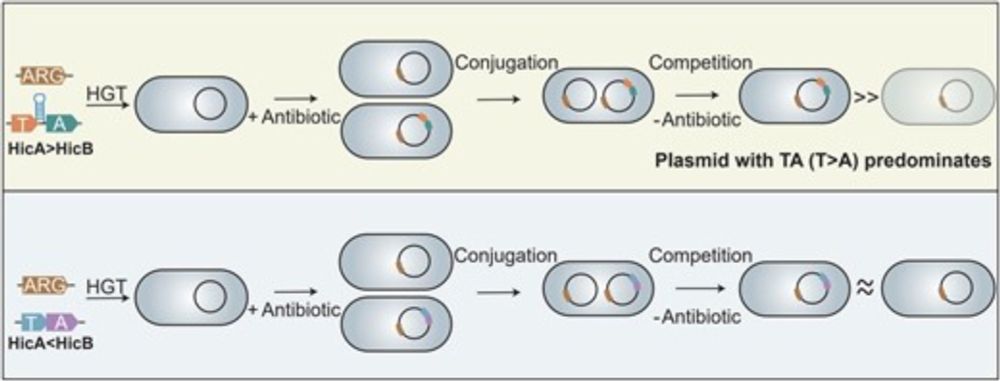
A noncanonical intrinsic terminator in the HicAB toxin–antitoxin operon promotes the transmission of conjugative antibiotic resistance plasmids
in @narjournal.bsky.social by Lin et al.
academic.oup.com/nar/article/...