Our new preprint is out@bioRxiv: www.biorxiv.org/content/10.1...
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
07.03.2025 00:37 — 👍 105 🔁 37 💬 3 📌 5

Red and blue poster supporting Stand Up for Science event 3-7 at Seattle Center noon to 3 includes a blue statue of liberty, and more information and a QR code. standupforscience2025.org
Among speakers Fri in #seattle,
brain surgeon Jim Olson, MD.
Who helped develop a "tumor paint" from scorpion 🦂
venom with @fredhutch.bsky.social
And @meadekrosby.bsky.social Climate scientist 🌏
Watch thread for more speakers @sufsseattle.bsky.social
🧵
04.03.2025 21:32 — 👍 18 🔁 8 💬 1 📌 0
Postbac Program
One of the many, many “pauses” and cancellations devastating our next generation of researchers. “The NIH Intramural Program has paused the recruitment of IRTAs, CRTAs, and Visiting Fellows in all training programs pending guidance from Health and Human Services. Check back daily for updates.”
18.02.2025 02:03 — 👍 123 🔁 48 💬 9 📌 17
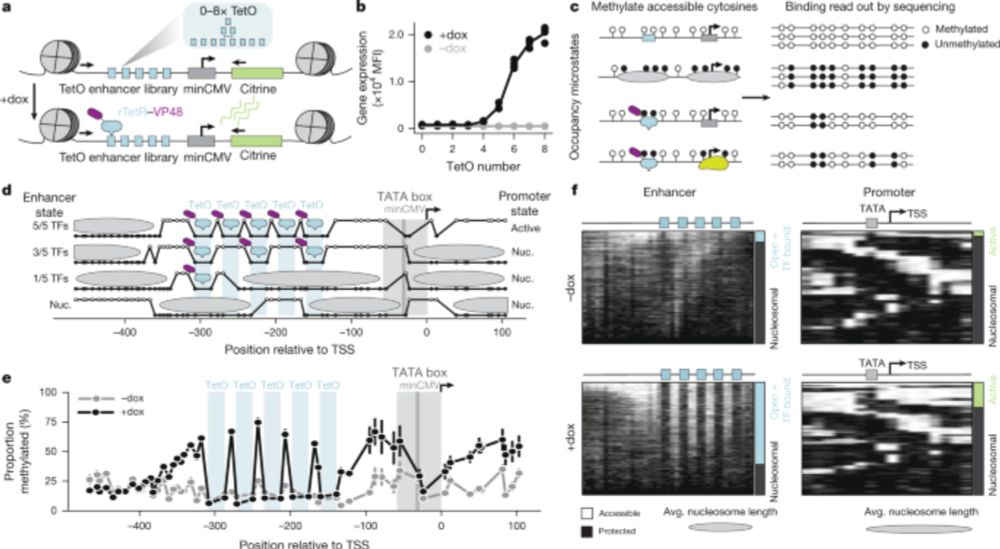
Chromatin conformation, gene transcription, and nucleosome remodeling as an emergent system
Gene transcription is an emergent phenomenon of packing domain geometry in situ.
In effect, our genomes are a very sophisticated computer system. We take measurements of the state (salts, food, stressors), integrate it with our prior measurements (chromatin remodeling from prior exposures, epigenetic memories of our grandparents) and adapt.
www.science.org/doi/10.1126/...
06.02.2025 14:53 — 👍 1 🔁 1 💬 1 📌 0
Let's start a conversation about this. I keep thinking about how what we do now spans across generations. #Epigenetics and #chromatin is a system for our genomes to tell our cells what to expect in the future and to prepare our children/grandchildren for current-ish conditions.
06.02.2025 14:53 — 👍 3 🔁 1 💬 1 📌 1
PREPRINT! Park et al. makes the case that we may be misunderstanding heterochromatin for past 30 years due to ChIP-Seq biases... 1/n
05.02.2025 18:34 — 👍 66 🔁 30 💬 5 📌 2
Frontiers | Inclusive mentorship of pediatric trainees: pediatric oncology as a microcosm
Very pleased - on this week of all weeks - to have this paper on the importance of inclusivity in mentorship published. Co-authored with the fantastic Drs. Sadhana Jackson and Jessica Tsai (@jestsai.bsky.social), we do a deep dive on pediatric oncology, but 1/3
www.frontiersin.org/journals/onc...
31.01.2025 20:44 — 👍 7 🔁 3 💬 1 📌 1
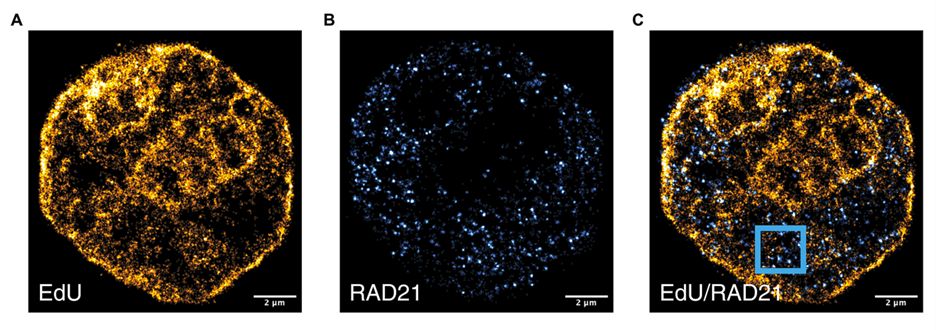
We hypothesized that this unique cohesin-mediated structural change in nascent chromatin domains may be caused by cohesin activity on the surface of mature chromatin domains. Using SMLM to image RAD21 and EDU-labeled chromatin, we found RAD21 primarily localized in DNA-poor areas between domains.
03.02.2025 16:15 — 👍 1 🔁 1 💬 1 📌 0
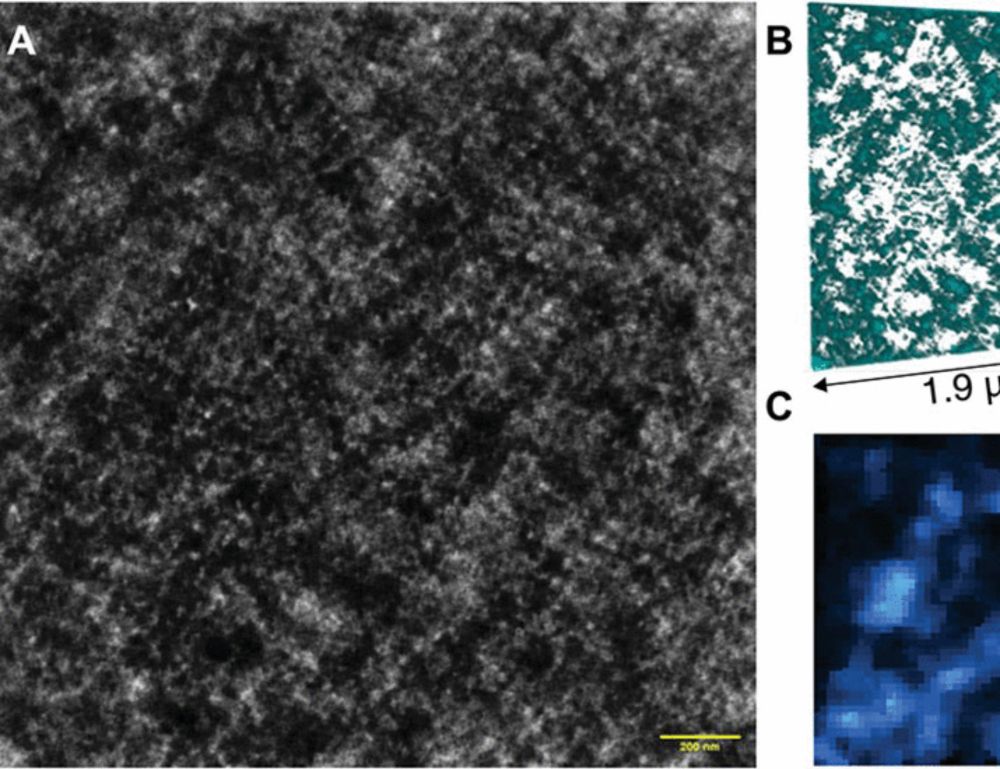
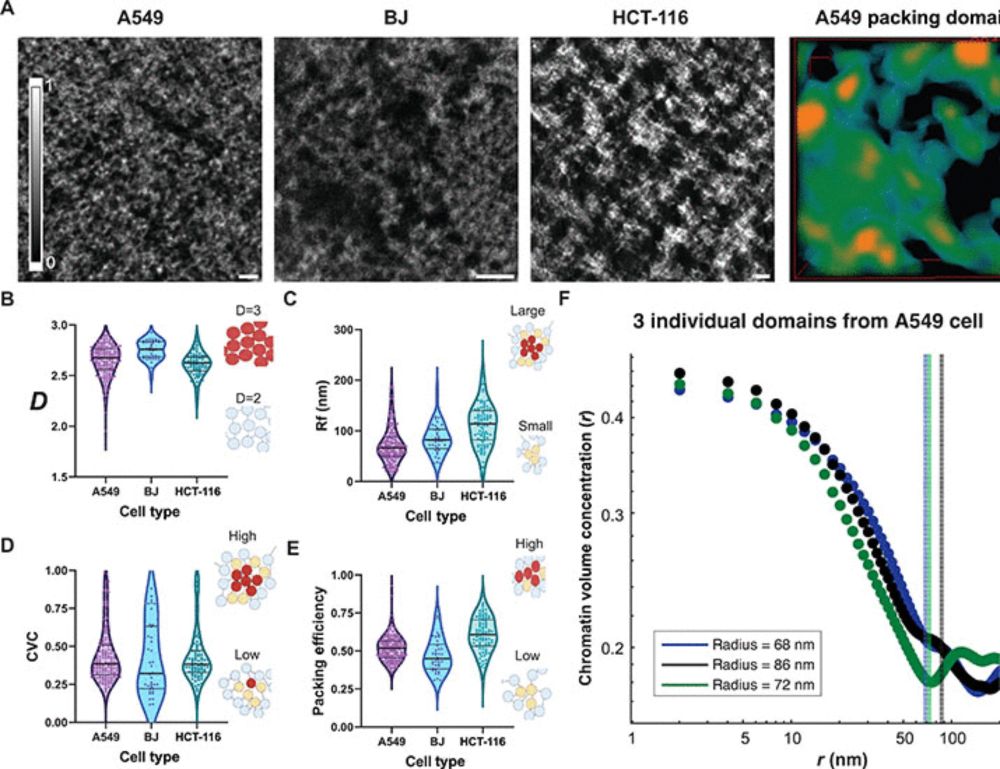
Mature chromatin packing domains persist after RAD21 depletion in 3D
RAD21 contributes to nascent packing domain formation, but maturation requires nucleosome posttranslational modifications.
CPGE's most recent publication on cohesin function and chromatin domain organization is out now in Science Advances. This thread will take a look at some of the key observations from the paper!
#chromatin #genomics #epigenetics #3Dgenome #northwestern #dna #cpge
www.science.org/doi/10.1126/...
03.02.2025 16:15 — 👍 9 🔁 2 💬 1 📌 1
Our paper on cohesin regulation of mesoscale chromatin organization is out
25.01.2025 00:57 — 👍 5 🔁 3 💬 0 📌 0
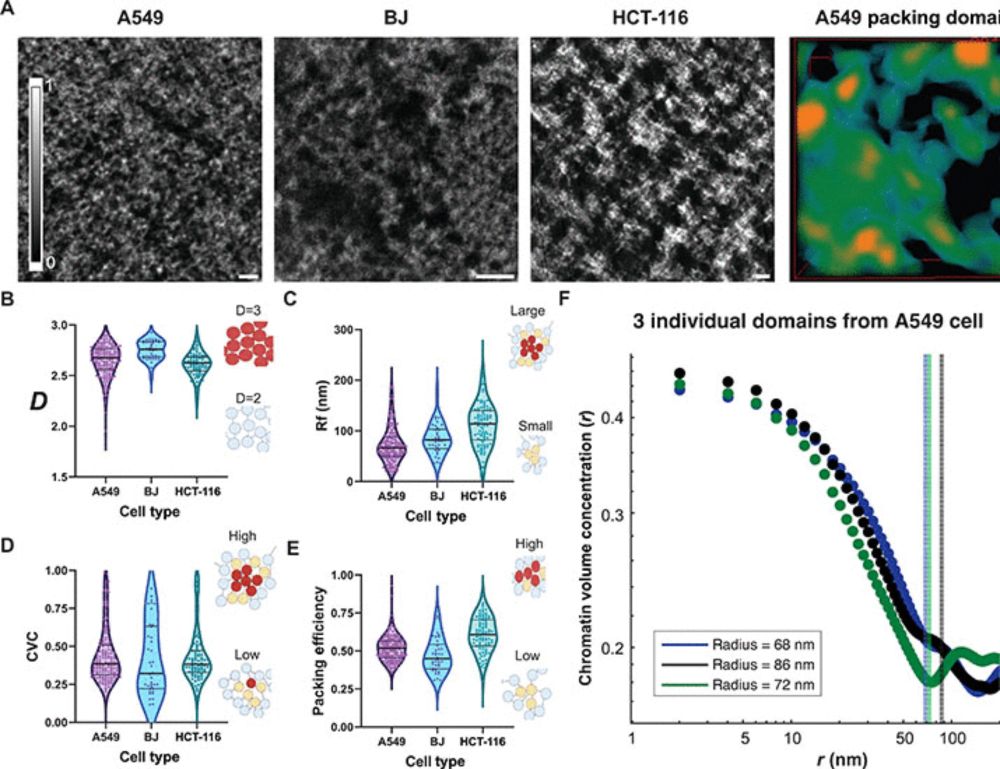
Chromatin conformation, gene transcription, and nucleosome remodeling as an emergent system
Gene transcription is an emergent phenomenon of packing domain geometry in situ.
There’s a lot to unpack in our recent Science Advances paper - come check it out! Here’s a thread on what we found.
#chromatin #genomics #biophotonics #dna #epigenetics #imaging #northwestern #science #biology #aging #longevity #biomedicine #cpge
www.science.org/doi/10.1126/...
14.01.2025 17:20 — 👍 19 🔁 11 💬 1 📌 0
Our latest paper on chromatin domains and genome function is out!
10.01.2025 20:13 — 👍 3 🔁 0 💬 0 📌 0
Hi Bluesky! I made a 3D chromatin starter pack.
Let me know if you would like to be added.
go.bsky.app/6tTQdqQ
13.11.2024 17:55 — 👍 84 🔁 42 💬 41 📌 0
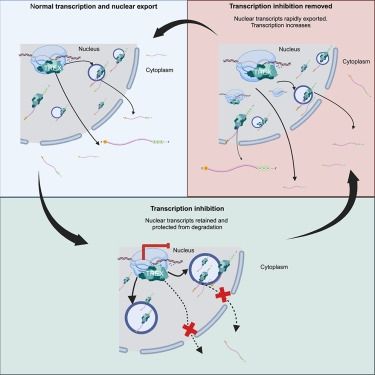
Out here at the #4DNucleome meeting in San Diego with a poster on our recent preprint where we explore the impact of Pol 1 and Pol 2 transcriptional inhibition on mesoscale chromatin organization.
#3DGenome
13.12.2024 14:41 — 👍 6 🔁 1 💬 1 📌 1
Center for Physical Genomics and Engineering
The mission of the Center for Physical Genomics and Engineering at Northwestern University's McCormick School of Engineering is to create new strategies for the treatment of disease and the reversible...
CPGE's archive of recorded talks can be viewed on our YouTube channel at www.youtube.com/@PhysGenCenter.
Visit us at www.physicalgenomics.northwestern.edu for details on our transdisciplinary research platform, training programs, and initiatives such as the Monthly Seminar on Physical Genomics!
12.12.2024 04:23 — 👍 1 🔁 1 💬 0 📌 0
Part of Wellcome Sanger Institute and funded by Wellcome, we develop and deliver genomics-focused learning and training. Our events support scientific careers, as well as the equitable application of genomics into research and healthcare across the globe.
Exploring the intersection of global health metrics, epidemiology, and data science. Bridging the gap between methods and practice to better measure and improve population health worldwide.
Associate Professor of Genetics at Washington University in St. Louis. I write about genomics at https://www.thisgenomiclife.org
Medicinal chemistry lab @IKCU, Faculty of Pharmacy, Izmir.
Drug discovery studies; design and synthesis new compounds as antileishmanial, antibacterial, anticancer, antiparasitic, FABPi, TYK2i
@istanbulluh.bsky.social
@huseyinkosker.bsky.social
Bluesky shadowbanned me
Follow my alt account
Current yeast articles everyday :)
Follow our Feed #yeast #saccharomyces
:)
From Molecular Genetics Lab, USACh
The Northwestern University Clinical and Translational Sciences (NUCATS) Institute is an integral link in Northwestern’s clinical & translational research enterprise, accelerating innovation by providing scientists with consultative resources and expertise
The CWP is a workers’ cooperative developing theory, analysis, and practice around issues relating to class, oppression, and stigma in Britain and beyond.
Publishers of Lumpen: Journal of Poor and Working Class Writers
https://linktr.ee/lumpen_journal
Working Class Environmentalism & Movement Building.
Founder and Co-Director of the Working Class Climate Alliance @wccalliance.org
https://linktr.ee/the_wcca
MPhil/PhD Social Anthropology at Goldsmiths University
Research and development company leveraging the biology of diverse organisms. Read about our science: http://research.arcadiascience.com
Co-founder & CSO @ArcadiaScience. Head of Open Science @AsteraInstitute. Immigrant. she/her
Still post frequently on X (@PracheeAC)
we highlight the latest articles that bring together physics and biology.
(maintained by @shuzokato.bsky.social)
Genseq provides ISO 15189 accredited clinical genetic testing services to healthcare professionals and a comprehensive range of genomic solutions to the biopharma and clinical research sectors worldwide.
Follow us for #genetics educational insights!
Postdoc/ PhD @kazu-maeshima.bsky.social at National Institute of Genetics, Japan / Chromatin, live-cell imaging, microscopy, recently interested in mechanobiology and cell differentiation
https://linktr.ee/shiori_iida
Group Leader (RyC) at IBFG in Salamanca, Spain @CSIC.es @usaloficial.bsky.social
Fascinated about protein PTMs and how they regulate life
Cell Signalling - Chromatin - PTMs
ERC starting #PTMtalk
Associate Professor @Cornell #Genetics #Epigenetics #Chromatin #Zebrafish #Development #Reproduction #EvoDevo #Bioinformatics #ZGA #StemCells #Regeneration #GoBills #BillsMafia 🦬🏈💪
Molecular biology lab based at the Department of Biology, University of Oxford. Researching insect epigenetics and epigenetic inheritance. Actively recruiting!
#Hospital, #MD, #Rheumatology, UK based.
A chemistry PhD & pharma bg, came to medicine late
Scrolling is a distraction from taking action. Pick some concrete way to help & get to it.
@twofishnocat.bsky.social for the likes/hobbies stuff
Group Leader, Genome Institute of Singapore, A*STAR, Working on computational methods for long read RNA-Seq
https://github.com/goekelab