The “Ins and Outs and What-Abouts” of H2A.Z: A tribute to C. David Allis - Journal of Biological Chemistry www.jbc.org/article/S002...
17.07.2025 16:03 — 👍 3 🔁 1 💬 0 📌 0

Check out the new chalk art outside the lab at Cornell!
11.07.2025 16:56 — 👍 7 🔁 0 💬 0 📌 0
@zebrafishrock.bsky.social could you please broadcast this?
08.07.2025 20:18 — 👍 1 🔁 0 💬 0 📌 0


Congrats Shan. It was wonderful to participate in the graduation ceremony with you! @urochester.bsky.social
22.05.2025 00:48 — 👍 4 🔁 0 💬 0 📌 0
Great event. Great trainees. Thank you!
30.04.2025 12:21 — 👍 2 🔁 1 💬 1 📌 0

Such an awesome day! It was wonderful to host Marisa Bartolomei @marisa-bartolomei.bsky.social as the Keynote speaker our annual Genetics Day symposium.
29.04.2025 20:51 — 👍 2 🔁 0 💬 0 📌 1
Congrats!!
05.04.2025 00:25 — 👍 2 🔁 0 💬 0 📌 0

Having an awesome time at the Tri-Repro meeting @pennmedicine.bsky.social @pennepigenetics.bsky.social with my good bud @themodzlab.bsky.social
04.04.2025 01:48 — 👍 6 🔁 0 💬 0 📌 0
I had a hypothesis that vitamin D prevents cancer. I worked on this for > 10 years, had 3 NIH-funded grants, and published >30 papers on this topic. Based on our work and that of my esteemed colleagues, I concluded that the evidence does not support the hypothesis.
RFK Jr., we are not the same.
28.03.2025 15:57 — 👍 2014 🔁 327 💬 31 📌 14
Had a wonderful time visiting the Genetics Department at the University of Georgia. Great food! Great conversation! Great company! and Excellent weather! Thanks for the invite @gollmg.bsky.social
28.02.2025 00:56 — 👍 1 🔁 0 💬 0 📌 0
A cropped screenshot of the Flashes app in the App Store
Congratulations to @flashes.blue for their official launch on the App Store 🎉🎉🎉
It’s an instagram alternative built on the AT protocol 🙌 all the visual scrolling with none of the Meta algorithm
25.02.2025 07:33 — 👍 10968 🔁 1839 💬 307 📌 220

Congrats Brandon! So proud of your accomplishments.
20.02.2025 18:43 — 👍 4 🔁 0 💬 0 📌 0

We all know the story of Henrietta Lacks, but there are countless other examples when biomedical research advanced with a little regard for morals or ethics - often at the expense of poor, disadvantaged, or minority individuals. This book has been enlightening. A good read for current scientists.
12.02.2025 14:35 — 👍 2 🔁 1 💬 0 📌 0
CONGRATS to all!!
11.02.2025 17:31 — 👍 5 🔁 1 💬 0 📌 0
Now that I have read this paper, my lab has new findings based on CUT&Tag data that challenge the traditional view of euchromatic and repressive factors. I have started doubting all previous ChIPseq-based conclusions for many targets we are interested in. Thanks for publishing the preprint
06.02.2025 17:55 — 👍 3 🔁 1 💬 0 📌 0
CUT&Tag Identifies Repetitive Genomic Loci that are Excluded from ChIP Assays https://www.biorxiv.org/content/10.1101/2025.02.03.636299v1
05.02.2025 16:38 — 👍 19 🔁 5 💬 1 📌 2
Very interesting another reason to stop doing Chip seq and do cut&tag. Looking forward to reading it
05.02.2025 20:57 — 👍 11 🔁 3 💬 0 📌 0
Thanks so much Ed!!
05.02.2025 19:50 — 👍 0 🔁 0 💬 0 📌 0
... Finally, Brandon purified proteins that would otherwise be lost in the pellet in ChIP-Seq, and identified several interesting factors, such as BRD4, NSD2, and RUNX1, (which co-precipitated with repetitive heterochromatin) suggesting these factors might bind and regulate silenced chromatin... 5/5
05.02.2025 18:37 — 👍 2 🔁 1 💬 0 📌 0
... Brandon then demonstrated that in situ chromatin fragmentation techniques, like CUT&Tag or CUT&Run, can overcome this problem, allowing him to characterize H3K9me3 enrichment over evolutionarily young repetitive elements.... 4/n
05.02.2025 18:34 — 👍 0 🔁 0 💬 1 📌 0
... He goes on to demonstrate that these biases are due to heterochromatin begin resistant to sonication, relegating some genomic loci to the insoluble pellet, meaning a good amount of heterochromatin cannot be pulled down w/ antibodies... similar to prior results from Ken Zaret's lab... 3/n
05.02.2025 18:34 — 👍 2 🔁 0 💬 1 📌 0
... Brandon, as shared student between my lab and @mitchelloconnell.bsky.social, has identified major biases in ChIP-Seq datasets, which prohibit the study of heterochromatic loci and repetitive elements in several mammalian cell types... 2/n
05.02.2025 18:34 — 👍 1 🔁 0 💬 1 📌 0
PREPRINT! Park et al. makes the case that we may be misunderstanding heterochromatin for past 30 years due to ChIP-Seq biases... 1/n
05.02.2025 18:34 — 👍 66 🔁 30 💬 5 📌 2
 04.02.2025 18:42 — 👍 5199 🔁 1554 💬 83 📌 48
04.02.2025 18:42 — 👍 5199 🔁 1554 💬 83 📌 48
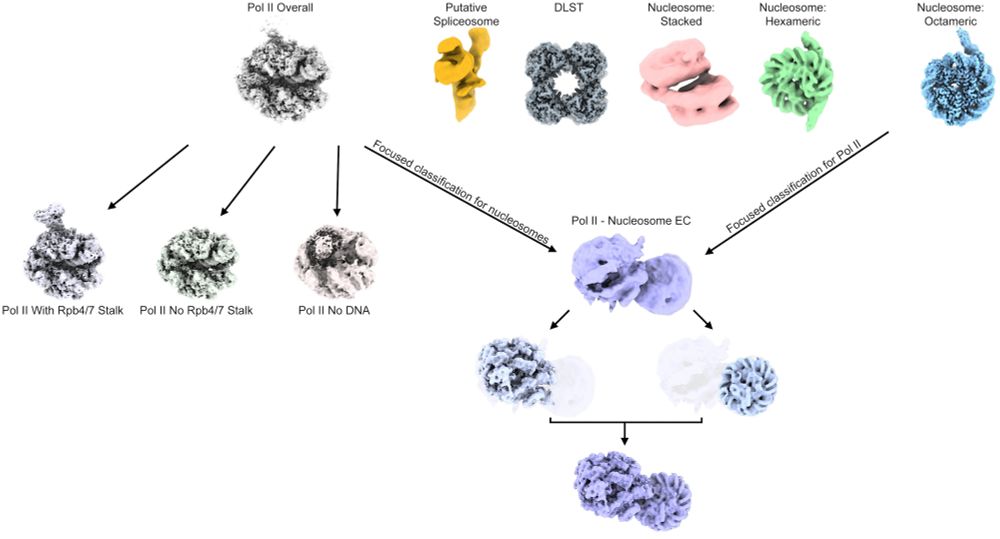
We did a thing... We purified native transcriptional complexes from Drosophila embryos, obtaining +/- stalk Pol II elongation complexes, native nucleosomes, as well as a nucleosome elongation complex. For more details please check out: doi.org/10.1101/2025...
05.02.2025 01:02 — 👍 56 🔁 20 💬 1 📌 1
Here’s why we need to study & fund developmental biology: to understand, prevent & treat birth defects. Infants can’t speak, so we must be their advocates
04.02.2025 16:35 — 👍 19 🔁 10 💬 0 📌 0
WTF!!! This is a for real disaster!! He needs to be stopped. We can’t have this anti-science lunatic leading HHS.
05.02.2025 01:30 — 👍 0 🔁 0 💬 0 📌 0
🤦♂️ shit.
05.02.2025 01:02 — 👍 1 🔁 0 💬 0 📌 0
HHMI Hanna Gray fellow & postdoc with Andy Clark and Cedric Feschotte at Cornell peiweichen.com
Studying principles of tissue regeneration in the axolotl and looking toward regenerative therapies. Asst. Professor at Wake Forest University. Views my own. He/him.
https://www.currie-regenerationlab.com/
Postdoc in Stainier Lab @ Max Planck Institute Bad Nauheim - gene expression regulation during embryogenesis - transcription and translation imaging in living embryos
Drosophila/zebrafish
Biochemistry, Molecular, and Cell Biology(BMCB) Doctoral Student @Cornell
Art Museum
🖼️5,000 years of world art in Rochester, NY.
🏛️Visit Wed-Sun from 11 am-5 pm & Thurs from 11 am-9 pm.
Official Bluesky account for the University of Rochester
Thinkers, makers, researchers, performers, healers—pushing boundaries to Make the World #EverBetter
About Us: https://www.rochester.edu/about/
Watch: https://www.youtube.com/user/universityrochester
Genomic imprinting and epigenetics. I will fight for science!
Senior editor @naturesmb.bsky.social. Views and opinions are personal.
Serra Group at UCSD Physics: Nonlinear Dynamics & Physics of Living Systems.
http://www.mattiaserra.com
Prev. |@SchmidtFellows @Harvard, PhD @ETH_en
Developmental biologist. Postdoc at the Francis Crick Institute
UCLA Professor and HHMI Investigator interested in plant epigenetics and plant genome editing.
Cllr. AKC. FRSA. Marathon Runner. History. Philosophy. Politics. Theology.
Leading conference on Eukaryotic Gene Regulation & Functional Genomics in Latin America. Volunteer-run. Biennial. Next edition: Sep 29- Oct 3, 2025.
📍Puerto Varas, Chile 🇨🇱
🌐More info at molbiosystems.com
An independent, non-profit biomedical research institution in Salisbury Cove, Maine, improving human health through research, education, and ventures that transform discoveries into cures: mdibl.org
Assistant PI Department of Genetics at University of Georgia
Previous Hawley lab postdoc and Duke Cell Bio grad student. All my views are my own (she/her)
Wandering around the complexities of biological phenomena related to genome regulation and anything connected to it 🧬
A dedicated fan of smiling out loud!
Chromatin, Epigenetics and Transcriptional Regulation Network connecting scientists at the University of Edinburgh and beyond
Husband, dad, veteran, writer, and proud Midwesterner. 19th US Secretary of Transportation and former Mayor of South Bend.
A multi-disciplinary, gold #OA journal publishing cutting-edge methodological advances. @cellpress.bsky.social
Computational biologist. Studies how humans respond to RNA viruses.

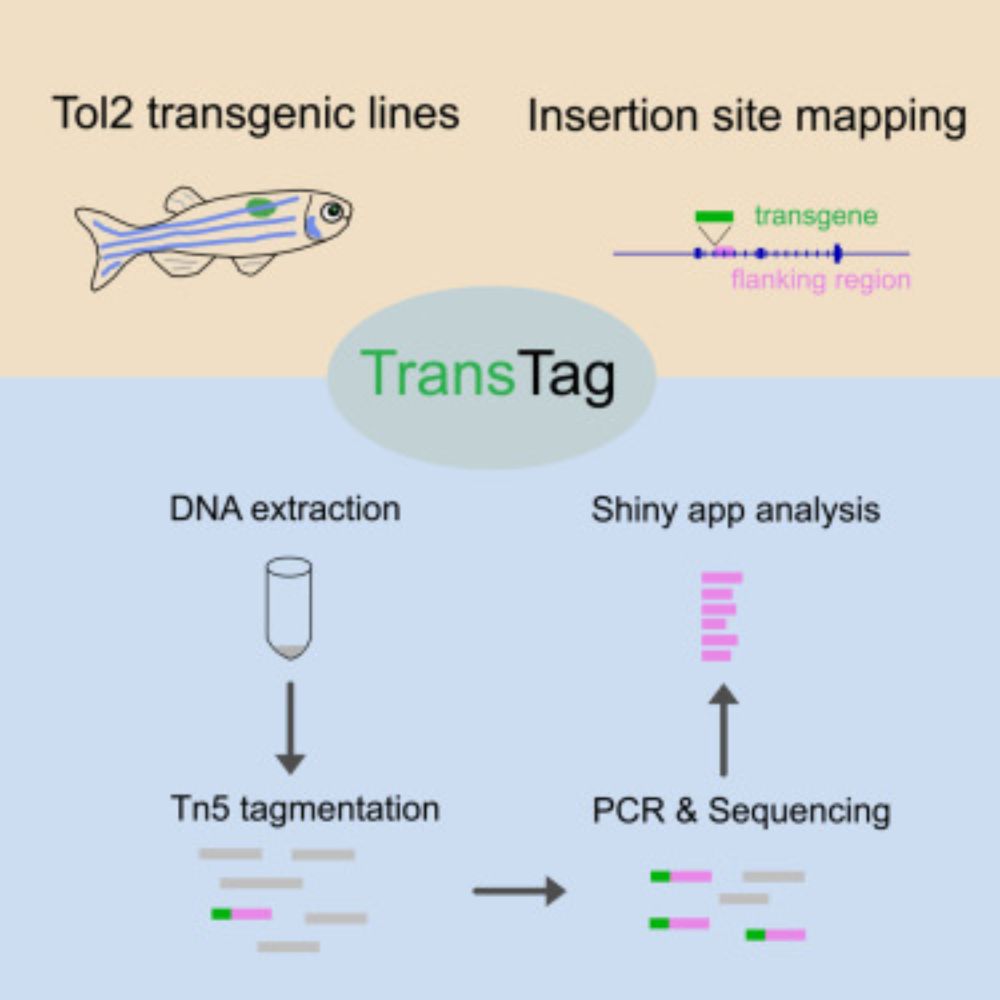







 04.02.2025 18:42 — 👍 5199 🔁 1554 💬 83 📌 48
04.02.2025 18:42 — 👍 5199 🔁 1554 💬 83 📌 48
