

📢📢📢 The #AsiaRNA2025 abstract deadline has been extended to September 5th!!!
Join us at the inaugural Asia RNA Club Symposium 2025
🗓️ Nov 3-5, 2025
📍Seoul National University, Seoul, 🇰🇷
Register by Oct 10th
More info: asiarnaclub.org
#RNASky @rnasociety.bsky.social
🔄🙏🏽
20.08.2025 09:47 — 👍 4 🔁 2 💬 0 📌 0
@selfdz.bsky.social @archafox.bsky.social @fukayalab.bsky.social
21.08.2025 08:04 — 👍 1 🔁 0 💬 0 📌 0

The Asia RNA Club Symposium will be happening in Seoul in Nov 3-5, bringing together RNA scientists from the Asia-Pacific region. Excellent speakers and a great opportunity to connect! Abstract submission by Sept 5 asiarnaclub.org
21.08.2025 08:04 — 👍 2 🔁 3 💬 1 📌 0

Narry Kim, Daniel MacArthur, Shi-Jie Chen, K Thangaraj, Ryan Lister, Karen Miga, Arnold Ou, Nikolaus Schultz, Leslie Beh, Koh Woon-Puay, Jingmei Li, PhD, Boxiang Liu, Jinyue Liu, Kristijan Ramadan, Patrick Tan, Yvonne Tay, Sunny Wong
🎉GIS turns 25! Join us in celebrating 25 years of innovation at #GIS25: Genome Innovation and Precision Medicine Conference, happening 11–12 September at Matrix, Biopolis, Singapore!
📅 Save the date and register now! Limited seats left!
👉 a-star.edu.sg/gis/news-eve...
#Genomics #PrecisionMedicine
20.05.2025 09:32 — 👍 4 🔁 1 💬 2 📌 0
I am not sure how to do Bluesky yet, but I want to tell the world about our neighborhood NMF method. First of many collaborations with the Pelka Lab!!
29.04.2025 23:35 — 👍 23 🔁 7 💬 0 📌 0

SINGAPORE SCIENTISTS UNVEIL ONE OF WORLD’S LARGEST LONG-READ RNA SEQUENCING DATASETS TO ADVANCE DISEASE RESEARCH
Scientists at A*STAR GIS have unveiled SG-NEx, one of the world's largest long-read RNA sequencing datasets! 🌍🔬 With 750M long RNA reads, it enhances detection of complex RNA features, aiding disease research and precision medicine. Available via AWS Open Data Registry. #Genomics #PrecisionMedicine
22.04.2025 09:48 — 👍 23 🔁 7 💬 1 📌 2
Join us at Genome Institute of Singapore! Great environment, stable funding!!
14.04.2025 05:26 — 👍 2 🔁 2 💬 0 📌 0
Excellent opportunity for PI positions at the Genome Institute of Singapore! Outstanding facilities, latest genomic technologies, strong and stable science funding. Junior candidates and established scientists are welcome to apply
14.04.2025 06:25 — 👍 12 🔁 11 💬 0 📌 0
Congratulations to @jonathangoeke.bsky.social, and our @astar-gis.bsky.social team on your SG-NEx project, providing a comprehensive resource that enables the development and benchmarking of computational methods for profiling complex transcriptional events at isoform-level resolution.
21.03.2025 03:14 — 👍 3 🔁 3 💬 0 📌 1

An Analysis presents benchmarking results from the Singapore Nanopore Expression consortium project, plus a valuable resource of datasets generated from various long-read and short-read RNA sequencing technologies.
@jonathangoeke.bsky.social @astar-gis.bsky.social
www.nature.com/articles/s41...
13.03.2025 18:36 — 👍 15 🔁 7 💬 0 📌 2
Delighted to see this published. Congratulations Ying Chen and @jonathangoeke.bsky.social
14.03.2025 00:00 — 👍 6 🔁 3 💬 0 📌 0
Dr Heng Li presented his renowned work on the challenges and promises of long-read sequencing at GIS. His presentation on de novo assembly and variant calling created much excitement and discussion onsite and online. View the recording here👉 shorturl.at/EkDOC
Photo credit: @jonathangoeke.bsky.social
13.03.2025 09:56 — 👍 4 🔁 2 💬 0 📌 0

Looking forward to the Long-Read Symposium tomorrow! I will be talking about long read RNA-Seq and the SG-NEx Project github.com/GoekeLab/sg-...
12.03.2025 09:43 — 👍 3 🔁 0 💬 0 📌 0
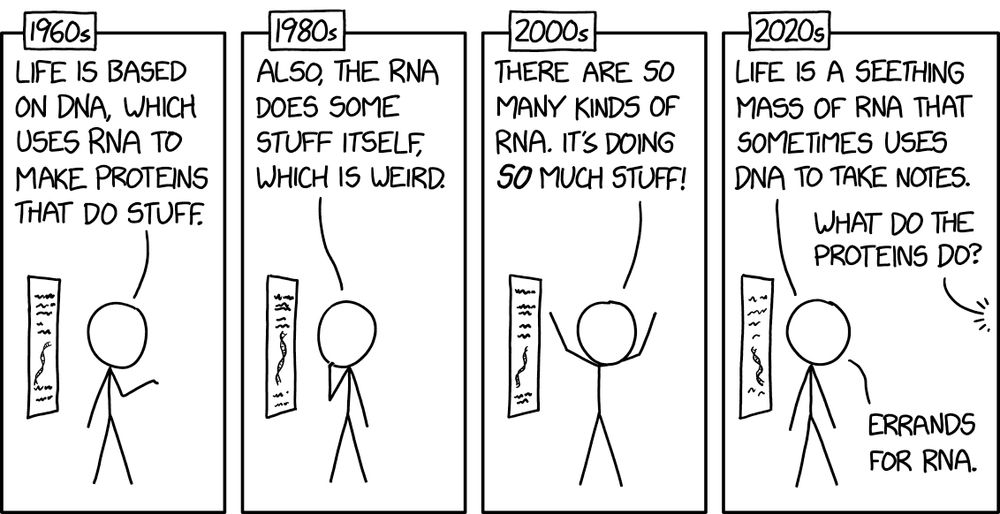
RNA xkcd.com/3056
26.02.2025 14:58 — 👍 17903 🔁 2568 💬 155 📌 171
Hi everyone! This is an amazing opportunity to join an incredible team! Great environment and interesting research! If you’re looking for a great place to grow, check this out and come join us!😊
23.02.2025 07:02 — 👍 6 🔁 4 💬 0 📌 0
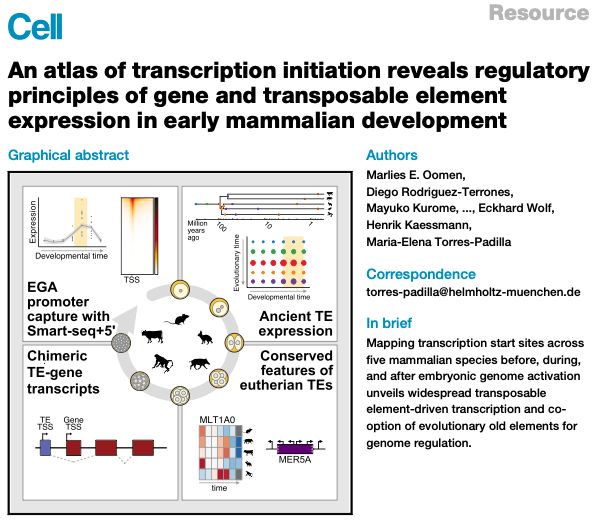
Paper out !!🥳big thanks to all authors @marliesoomen.bsky.social @diego-rt.bsky.social @kaessmannlab.bsky.social @jonathangoeke.bsky.social @lorenzamottes.bsky.social & 'bluesky-less'
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
21.02.2025 15:50 — 👍 143 🔁 53 💬 8 📌 2
Great opportunity, great lab setting global standards in the field of long read RNA! Highly recommended if you’re at this stage in your career
13.02.2025 11:39 — 👍 1 🔁 1 💬 0 📌 0

We are recruiting! We have an open position for a postdoctoral fellow to join our team at the Genome Institute of Singapore to work with long read RNA-Seq data. It’s a beautiful city state and outstanding research environment! More details here: jglab.org/postdoc-posi...
13.02.2025 09:38 — 👍 9 🔁 6 💬 0 📌 2
Our new paper examining how to analyse longread RNA-seq with no reference genome. We compare approaches for assembly and downstream analysis, from transcript accuracy to differential expression. Lead by @alexyfyf.bsky.social. Thnx to all contributors incl. @qgouil.bsky.social for the pea data!
10.02.2025 00:39 — 👍 28 🔁 11 💬 1 📌 0
(reposted with correct URL)
05.02.2025 08:47 — 👍 0 🔁 0 💬 0 📌 0

...reads are only assigned to transcripts after similar cells are clustered into groups, reducing the noise in transcript expression estimates due to missing data. We also provide unique counts for single cells (and EM counts for genes/experiments with high sequencing depth)
05.02.2025 08:47 — 👍 1 🔁 0 💬 1 📌 0
The challenge with transcript expression estimation is the limited number of reads per gene, which makes the assignment of (non unique) reads to transcripts difficult. To address this, Bambu performs a clustered EM, in which ....
05.02.2025 08:47 — 👍 1 🔁 0 💬 1 📌 0
PhD Student @ JHU Langmead Lab
Postdoctoral researcher at the Walter and Eliza Hall Institute in Matt Ritchie’s lab.
Research topic: Genomics | Transcriptomics | Long-read sequencing | Single-cell sequencing.
Principal Investigator at Genome Institute of Singapore, A*STAR
Laboratory of RNA Translation
Associate Professor of Biomolecular Engineering at the University of California, Santa Cruz; Associate Director, UC Santa Cruz Genomics Institute
Research group leader at the Max Planck Institute for Biology of Ageing in Cologne, Germany.
Excited by RNA-binding proteins, metabolism, stem cells and ageing research
Sr Research Fellow, Jay Shin Lab, Genome Inst Singapore. Collab Bruno Reversade Lab
PhD Eric Miska & Azim Surani Labs, Gurdon Inst, Cambridge
Biochem Grad, Oxford
Scientist interested in RNA and gene regulation. My passions are paraspeckles, RNA therapeutics for cancer and everything hiking/nature/community.
Institute for Quantitative Biosciences, UTokyo. We study the mechanism of transcriptional regulation in the Drosophila embryo.
https://sites.google.com/view/fukaya-lab-iqb/home
Genetics, transcriptomics, RNA and neuroscience.
Lab head at the University of Melbourne, Australia.
View own.
Bioinformatics and Computational Genomics at @HHU.de. Algorithms for pangenomes, structural variation, genome assembly, haplotype phasing, etc.
Statistical geneticist. Professor of Human Genetics and Biostatistics at the University of Pittsburgh. Assiduously meticulous.
Principal Bionformatician
@nanopore. Ex: Postdoctoral fellow @ NIH; Researcher @ CAB. Views are my own; #StandWithUkraine Support Ukraine!
Human Geneticist, Emmy Noether Group Leader at Helmholtz Munich & Dr. von Hauner Children‘s Hospital, University Hospital LMU Munich, Germany, Member of Die Junge Akademie
Director of Institute for Computational Genomic Medicine at Goethe University Frankfurt https://cgm.uni-frankfurt.de/
Professor, Investigator, mom, frequent flyer.
Group leader of Chromatin (Dys)function lab @MDC-BIMSB
Former Postdoc @Mundlos lab
PhD @ Schirmer lab
https://www.mdc-berlin.de/robson
A bioinformatics postdoc at Victoria Popic's lab at Broad Institute.
Molecular Biologist (aemonten.github.io) 丨Chief Editor @ CSH Protocols (cshprotocols.cshlp.org) 丨Head of the Integrity in Publishing Group at CSHL Press 丨Chair "Molecular Biosystems Conference" (molbiosystems.com) 丨(Oxford) Comma King 丨Central Dogma Police




















