Published in Science Advances! Looking forward to discussing the implications of our results and their applications with many researchers.
www.science.org/doi/10.1126/...
20.11.2025 02:24 — 👍 9 🔁 5 💬 1 📌 0

Gene-scale in vitro reconstitution reveals histone acetylation directly controls chromatin architecture
Reconstituting 20-kb chromatin shows that tuning acetylation alone reshapes its folding, dynamics, and contact domain formation.
To probe gene-scale chromatin physics, we built 96-mer (20 kb) arrays with defined histone marks. Combining single-molecule tracking, AFM imaging, and developing in vitro Hi-C, we saw how specific modifications dictate chromatin structure and dynamics. www.science.org/doi/10.1126/...
20.11.2025 07:46 — 👍 24 🔁 11 💬 2 📌 0
I launched my lab at UMass Amherst this fall. We study the mechanisms of meiotic homolog pairing using advanced live‑cell and super‑resolution imaging. Currently in budding yeast, with plans to expand to other eukaryotes. #Chromosome #Meiosis #Mitosis
www.umass.edu/biology/abou...
13.11.2025 12:13 — 👍 15 🔁 5 💬 1 📌 0
Welcome @shiori-iida.bsky.social , it’s great to have you in the lab! Excited about new directions in chromatin rheology and cell fate 🤓
20.10.2025 19:24 — 👍 6 🔁 1 💬 0 📌 0
I’ve recently joined @sarawickstrom.bsky.social lab at @mpi-muenster.bsky.social as a postdoc!
I’ll keep doing exciting science with joy and passion 🔬💫
Huge thanks to @kazu-maeshima.bsky.social lab for all the support during my PhD journey⛰️
I’ll start by tasting every single sausage here🇩🇪
20.10.2025 18:43 — 👍 6 🔁 0 💬 0 📌 1
Is euchromatin really “open”? Our new study @bioRxiv suggests otherwise. Using super-resolution imaging @shiori-iida.bsky.social @masaashimazoe.bsky.social reveals: Euchromatin forms condensed domains in live cells. Cohesin constrains them and prevents domain mixing.
www.biorxiv.org/cgi/content/...
28.08.2025 04:18 — 👍 103 🔁 37 💬 3 📌 3
Euchromatin is mostly condensed⁉️
With @shiori-iida.bsky.social , We revealed the detailed structure of euchromatin using super-resolution imaging.
Cohesin regulates the function of euchromatin via chromatin dynamics!
(1/n)
28.08.2025 08:43 — 👍 16 🔁 3 💬 3 📌 0
Thank you, Amro! Grad to see you soon ☺️
30.08.2025 13:57 — 👍 1 🔁 0 💬 0 📌 0

You can see that the slower subpopulation localizes like heterochromatin, while the faster one resembles euchromatin.
29.08.2025 04:12 — 👍 4 🔁 0 💬 0 📌 0
One more movie!
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
29.08.2025 00:36 — 👍 45 🔁 16 💬 2 📌 0
Thank you! ☺️
29.08.2025 00:34 — 👍 1 🔁 0 💬 0 📌 0
I’m grateful to my supervisor
@kazu-maeshima.bsky.social
for leading this project, my amazing co–first author
@masaashimazoe.bsky.social
for putting this paper together, all coauthors, supporters, and lab members!
Check out our opinion article, too:
www.sciencedirect.com/science/arti...
28.08.2025 15:15 — 👍 0 🔁 0 💬 0 📌 0
Our new preprint is out!
We combined single-nucleosome imaging and 3D-SIM to reveal:
🔹 Euchromatin forms condensed domains, not open fibers
🔹Cohesin loss increases nucleosome mobility without decompaction
🔹Cohesin prevents neighboring domain mixing
Full story & movies👇
28.08.2025 15:11 — 👍 19 🔁 7 💬 3 📌 1

Conserved nucleocytoplasmic density homeostasis drives cellular organization across eukaryotes
Nature Communications - Cells can regulate their mass density. Here, the authors demonstrate how eukaryotes establish and maintain a lower density in the nucleus than in the cytoplasm via pressure...
FINALLY! Challenging to publish but we believe it is an important discovery: rdcu.be/eATFz
💚 Thanks to the team @biswashere.bsky.social, Omar Muñoz, ✨Q✨ C. Hoege, B. Lorton, R. Nikolay @matthewkraushar.bsky.social @dshechter.bsky.social @gucklab.bsky.social @vasilyzaburdaev.bsky.social 💚
15.08.2025 11:54 — 👍 55 🔁 25 💬 1 📌 5

My #PhD defense work is on #Biorxiv!
Link: biorxiv.org/content/10.110…
Using single-molecule #imaging, we showed that liquid-like BRD4-NUT condensates locally constrain #chromatin via #bromodomain-dependent crosslinking.
I thank my supervisor, Prof. @kazu-maeshima.bsky.social, for his support!
16.07.2025 14:21 — 👍 4 🔁 2 💬 0 📌 0
Our new preprint 🧬🔗 www.biorxiv.org/content/10.1...
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
10.07.2025 04:21 — 👍 51 🔁 26 💬 1 📌 0

📢 NIG Postdoc Fellowship Call!
www.nig.ac.jp/nig/career-d...
🧬We’re seeking motivated postdocs to join our lab. 🔬We study chromatin behavior in living cells using advanced live-cell imaging and quantitative analysis:
www.science.org/doi/10.1126/...
doi.org/10.2183/pjab...
Interested? Please apply!
30.04.2025 15:27 — 👍 13 🔁 11 💬 0 📌 0
Check out this exciting preprint from @aznarbenitahlab.bsky.social discovering a new mode of crosstalk between YAP and the circadian regulator BMAL1. Congrats @juliabonjoch.bsky.social and @guiomarsolanas.bsky.social for fantastic work and thank you for the fun collaboration!
23.04.2025 19:32 — 👍 9 🔁 3 💬 2 📌 0
In developing embryos, cells move a lot! Plenty of that movement is random. Is random cell mixing a feature or bug for tissue patterning? Turns out, it’s both! Excited to share the 1st preprint from my postdoc w/ Sean Megason @seanemcgeary.bsky.social and Allon Klein. 1/20 doi.org/10.1101/2025...
24.04.2025 13:18 — 👍 122 🔁 34 💬 7 📌 6
🧬My retrospective chromatin review is out @Proc Jpn Acad:
doi.org/10.2183/pjab...
The textbook view of #chromatin as 30-nm fibers is fading.
A new paradigm: chromatin forms #liquid-like domains—locally dynamic, yet globally stable at the chromosome level (viscoelastic)—supporting genome function. ✨
25.04.2025 00:24 — 👍 28 🔁 13 💬 1 📌 0
Our new paper is out@ScienceAdvances👇
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
29.03.2025 01:06 — 👍 70 🔁 25 💬 1 📌 2


On top of that, I’m honored to receive the SOKENDAI Award from my university for my PhD work!
I can't thank @kazu-maeshima.bsky.social, my labmates, and all the NIG members enough for their support.
This recognition gives me even more motivation for the journey ahead!
27.03.2025 02:34 — 👍 9 🔁 0 💬 0 📌 1

It took blood, sweat, tears, too much coffee☕️, and just the right amount of alcohol🍺—but I finally got my PhD!
Huge thanks to everyone supported me, especially my mentor @kazu-maeshima.bsky.social, labmates, friends, and family.
I’m excited to keep chasing big questions and growing as a scientist!
27.03.2025 02:19 — 👍 11 🔁 0 💬 0 📌 1
Our new preprint is out@bioRxiv: www.biorxiv.org/content/10.1...
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
07.03.2025 00:37 — 👍 116 🔁 40 💬 3 📌 7
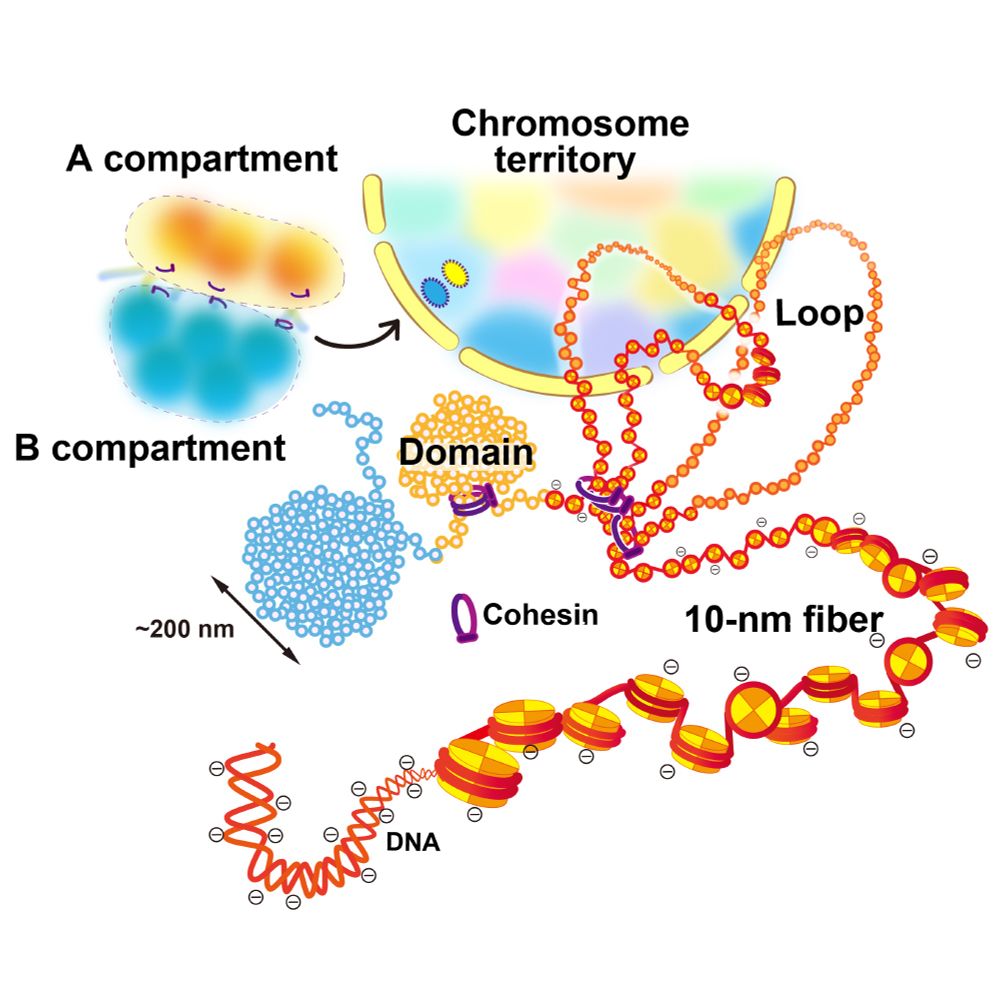
Our new review on how the #chromatin domain is formed in the cell is now available @Curr Opin Struct Biol.📄✨
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
20.02.2025 11:14 — 👍 75 🔁 37 💬 0 📌 0

Mature chromatin packing domains persist after RAD21 depletion in 3D
RAD21 contributes to nascent packing domain formation, but maturation requires nucleosome posttranslational modifications.
Today in Science Advances: "Mature chromatin packing domains persist after RAD21 depletion in 3D". Researchers at CPGE integrated measurements of genome connectivity and physical structure to deepen our understanding of chromatin organization
#chromatin #genomics
www.science.org/doi/10.1126/...
24.01.2025 22:32 — 👍 12 🔁 3 💬 0 📌 1

Thanks🎅🏼🦌
Merry Christmas all.
@shiori-iida.bsky.social @dai.tsujino
#NIGchrismtmass
24.12.2024 12:04 — 👍 6 🔁 2 💬 0 📌 0
We are seeking a motivated postdoc to join our lab! Our research focuses on live-cell imaging/analysis to reveal how chromatin behaves in living cells and how its behavior contributes to cellular functions.
www.nature.com/articles/s41...
www.science.org/doi/10.1126/...
Interested? Please DM me!
30.11.2024 12:32 — 👍 66 🔁 40 💬 2 📌 2
PhD student in the Bewersdorf Lab at Yale
Light microscopy | quantitative phase contrast imaging | super-resolution imaging | optical engineering
Associate Professor, University of Pennsylvania, Penn Epigenetics Institute
3D Genome structure and function
https://ericjoycelab.com/
Biologist @hubrechtinstitute.bsky.social interested in signalling dynamics, development, tissue homeostasis and microfluidics | Formerly MPI Biochemistry, Biocenter Basel and Embl Heidelberg | Mother of 2 | Posts are my own.
Utrecht | sonnenlab.org
President of the Max Planck Society since 2023 – dedicated to advancing science for the benefit of society. | @maxplanck.de
🔗 https://www.mpg.de/praesident
🔗 www.mpg.de/imprint
Group Leader @MPI-CBG and @POL, TU-Dresden. Dev Biologist mixing it up w/ Physics. Want to know how organs grow! also obsessed with structural colors | ritamateus.com
Group leader at MPI Molecular Biomedicine. Growing organoids and poking embryos to understand how functional organs are built.
https://www.mpi-muenster.mpg.de/researchgroups/rocha
Postdoc Wickström lab; PhD Stainier lab; @sheffielduni graduate. 👩🔬🧶🏋️♂️📚🐈🇷🇴
Interested in cell/dev bio and beautiful images🔬
We study the cell nucleus and the tissue microenvironment, driving dynamic cell mechanics. Multiscale Bioengineering Lab at College of Engineering & Mathematical Sciences at the University of Vermont. mccreerylab.com
Stem cell and mechanobiologist at the Max Planck Institute of Molecular Biomedicine and University of Helsinki
Bioengineer interested in #mechanobiology of cell state regulation, cancer, genome integrity, #chromatin & nuclear mechanics
RNAPII &Transcription Regulation -Chromatin Conformation & Nuclear Architecture - ESCs & Neuronal Differentiation - Nuclear Mechanotransduction - Cell Plasticity
UMG, Göttingen, Germany
Scientist of Living Matter. ICREA Professor at the Institute for Bioengineering of Catalonia, Barcelona. The need to know.
Confinement Mechanobiology Lab, Mechanobiology Institute | National University of Singapore
The Epigenetic Face of Cancer Metabolism Lab
We study NUCLEAR METABOLISM at CRG, Barcelona
https://sdelcilab.crg.eu/
Biomechanics, Developmental biology, Cell biology
PhD student at Nano-LSI, Kanazawa Univ. (D1)
#Mechanobiology #DevelopmentalBiology #Biophysics
Japanese Society of Developmental Biologists: exciting events/papers will be shared.
日本発生生物学会公式:関連イベントやおすすめ論文などを発信します
IMPRS-GS PhD Student at Papantonis Lab, University Medical Center Göttingen | BSc. Biochemistry and Biotechnology, UTH | MSc. Stem Cells and Regenerative Medicine, AUTH
Research Scientist at Institut Curie.
Postdoc at Tjian/Darzacq Lab, UC Berkeley.
PhD at Ellenberg Lab, EMBL Heidelberg.
Previously at Boutros Lab, Pines Lab, Stemmann Lab.
LinkedIn: nike-walther-836252b5
Research Scientist at Haruhiko Koseki lab. at RIKEN IMS🇯🇵 My research has focused on limb development, Polycomb group proteins, Hox and chromatin organization🐓🐀🧬 Opinions are my own




















