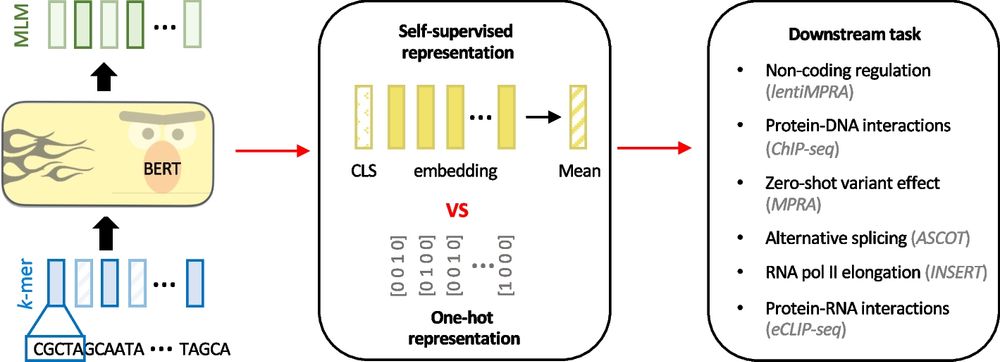
Excited to be at #ASHG2025 in Boston next week. On Thursday, I will be presenting a poster (1007T) on our latest work on Personalised sequence to expression modelling. Looking forward to meeting new folks and reconnecting with old friends and colleagues.
10.10.2025 02:49 — 👍 11 🔁 2 💬 0 📌 1

My team has a postdoc position available now, join us!
careers.gene.com/us/en/job/20...
19.09.2025 20:12 — 👍 1 🔁 4 💬 0 📌 1

Excited to be at @aithyra.bsky.social #AIinLifeScience symposium in Vienna. What a gorgeous venue!
Will be presenting posters on my latest work on personalized gene expression prediction and gene regulatory network inference
08.09.2025 15:17 — 👍 3 🔁 0 💬 0 📌 0
Dived into past, present & future of human genetics with brilliant students & mentors.Grateful for the chance to present my work on personalized sequence→expression prediction and discussions with @sashagusevposts.bsky.social @bpasaniuc.bsky.social @mashaals.bsky.social @tuuliel.bsky.social & others
01.08.2025 18:07 — 👍 9 🔁 1 💬 0 📌 0
All the best @paubadiam.bsky.social 🎉🎉
23.06.2025 11:46 — 👍 1 🔁 0 💬 0 📌 0
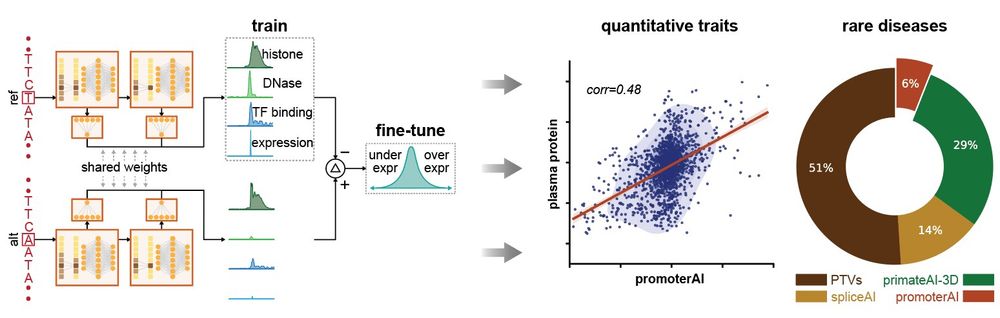
Excited to share my first contribution here at Illumina! We developed PromoterAI, a deep neural network that accurately identifies non-coding promoter variants that disrupt gene expression.🧵 (1/)
29.05.2025 23:57 — 👍 60 🔁 21 💬 1 📌 1
Thanks for your thoughts
22.05.2025 12:33 — 👍 0 🔁 0 💬 0 📌 0
I am genuinely wondering and curious what you think. It’s increasingly hard to see how academia can attract or retain top talent with offers like this, especially when industry offers 2–3x more. Something structural has to change if we’re serious about advancing AI in science.
22.05.2025 09:20 — 👍 1 🔁 0 💬 1 📌 0
Hi Andrea,
Thanks for sharing this, it’s an exciting and meaningful opportunity. But to be honest, the listed salary (€42–49k for postdocs) is deeply misaligned with the qualifications you're asking for- post-PhD experience, publications in top AI and biology journals, teaching, and vision.
22.05.2025 09:20 — 👍 1 🔁 0 💬 0 📌 0
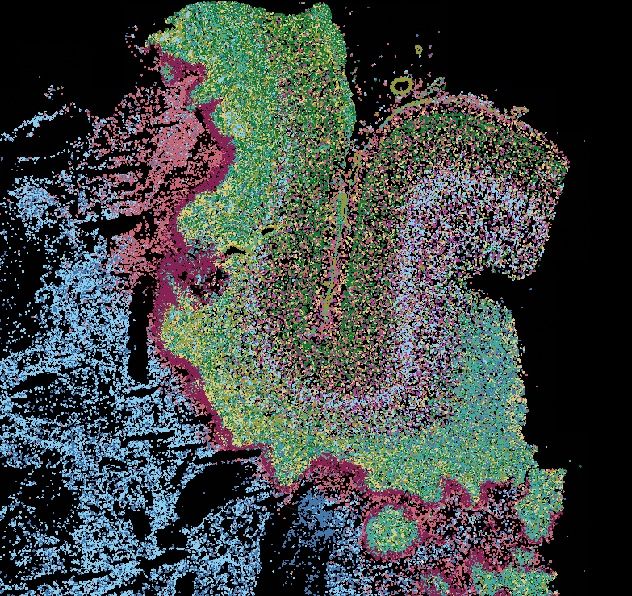
How does tumour heterogeneity arise? How can we predict cancer cell plasticity? In 2 new studies, we trace #glioblastoma heterogeneity to a spatial cancer cell trajectory w. multimodal cell atlassing bit.ly/4mkrWgs & predict plasticity w. snRNA/ATAC+deep learning bit.ly/3FbI6Ic 🧵
16.05.2025 11:42 — 👍 73 🔁 31 💬 6 📌 5
Thanks a lot for the shoutout Stein. scDORI builds upon the insights from the foundational works in GRN inference from your lab - Cistopic, SCENIC, SCENIC+
Thanks a lot for your contributions. Very exciting time to be in the field 🚀
16.05.2025 10:10 — 👍 2 🔁 0 💬 0 📌 0
Thanks to all our collaborators, specially @lauraruedag.bsky.social who co-led the computational analysis and was the best partner in crime one could ask for. What a delight it was!
Thanks to Elisa, Tannia and Fani for leading the experimental aspects.
16.05.2025 10:04 — 👍 4 🔁 1 💬 1 📌 0
14/ We believe this approach can be applied to other cancers to uncover—and exploit—plasticity brakes. Get in touch if interested! #GBM #MultiOmics #CancerResearch #deeplearning #cancerneuroscience #GRNs #Cancer #singlecell
16.05.2025 10:04 — 👍 2 🔁 0 💬 1 📌 0
13/ Our framework unifies regulatory and spatial logic of GB heterogeneity. While our companion paper showed subclones are intermixed across conserved tissue niches, our regulatory model explains WHY—they follow the same trajectory because they're constrained by the same regulatory rules!
16.05.2025 10:04 — 👍 1 🔁 0 💬 1 📌 0
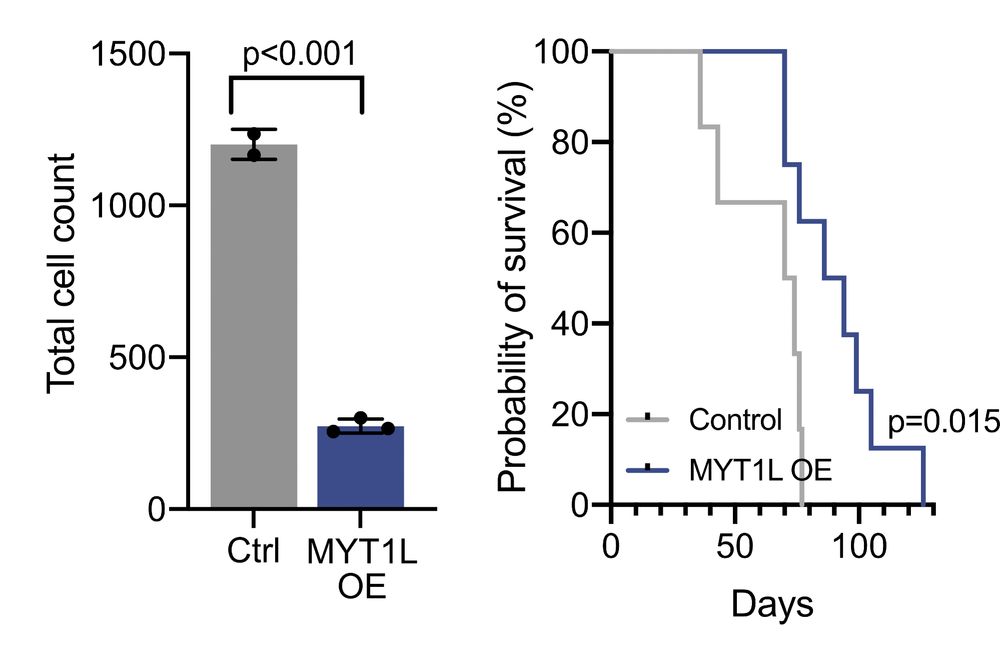
12/ Beyond chromatin changes, MYT1L transformed GB cells into less aggressive neuronal-like cells with: • Enhanced neurite-like morphology • Reduced tumor microtube connectivity • Decreased proliferation • In vivo: slower growth, less invasion, longer survival!
16.05.2025 10:04 — 👍 1 🔁 0 💬 1 📌 0
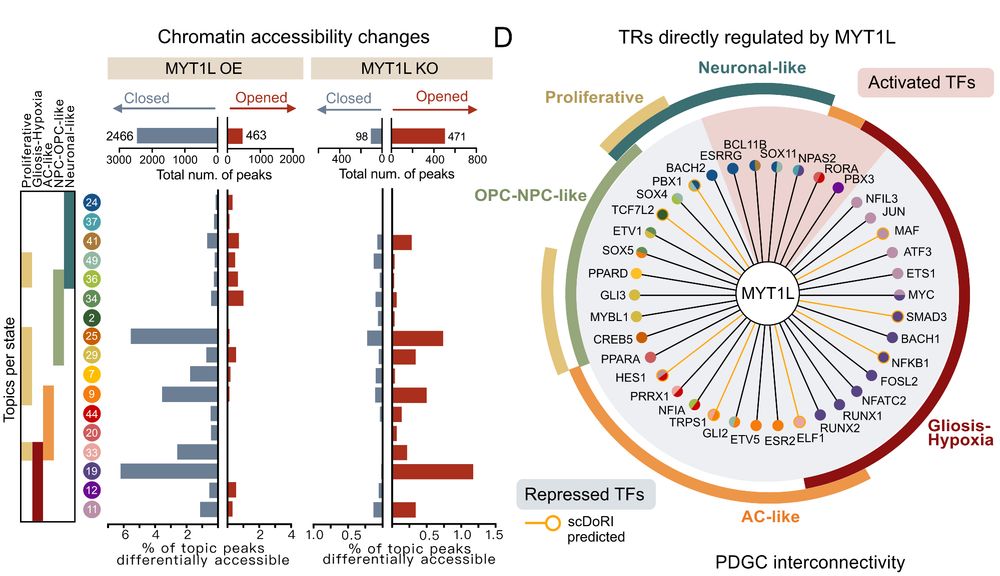
11/ Our experimental validation was striking: • MYT1L overexpression closed >80% of differential chromatin regions • MYT1L knockout reopened access to plastic fates • MYT1L directly bound and repressed regulators of other GB states• 85% of scDORI's predicted TF targets confirmed!
16.05.2025 10:04 — 👍 1 🔁 0 💬 1 📌 0
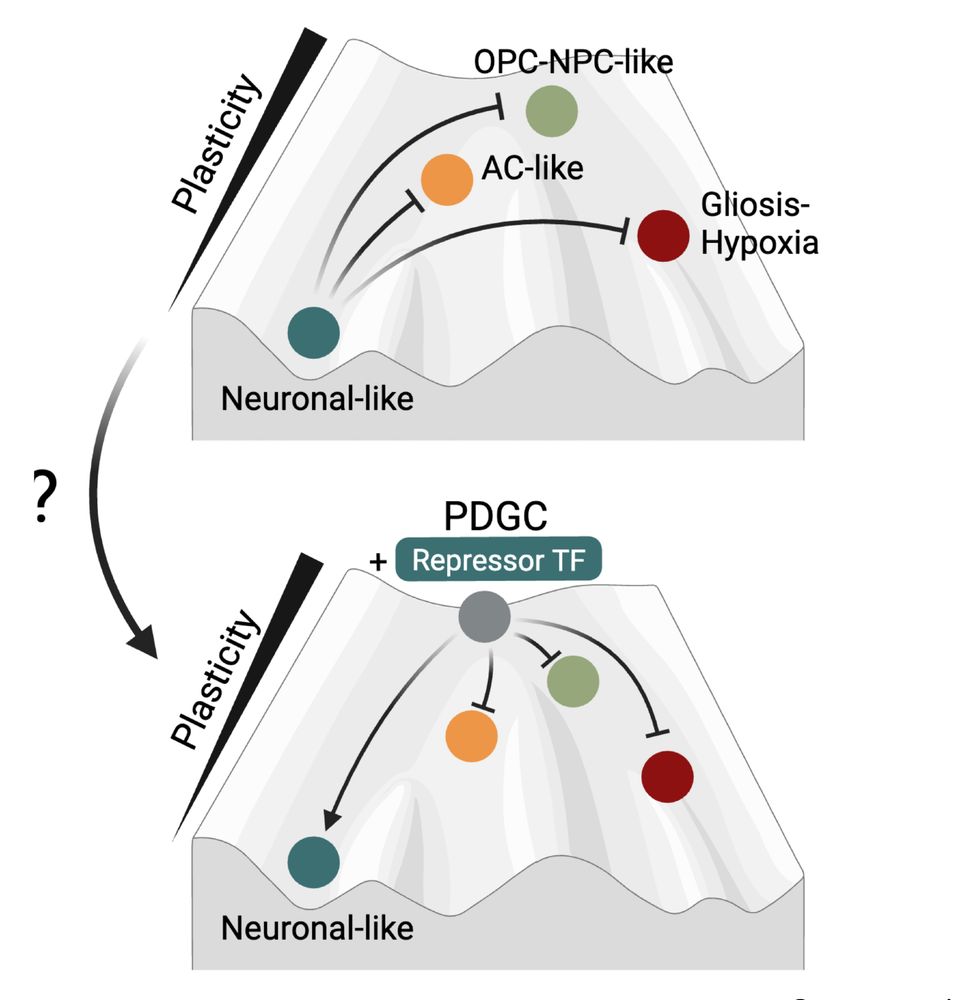
10/ 🔥 Can we use these GRNs to manipulate tumor identity and push GB cells into less plastic states? YES! We predicted MYT1L as the key regulatory bottleneck—a master repressor that locks cells into neuronal-like states by directly binding and suppressing the regulators of plastic states.
16.05.2025 10:04 — 👍 1 🔁 0 💬 1 📌 0
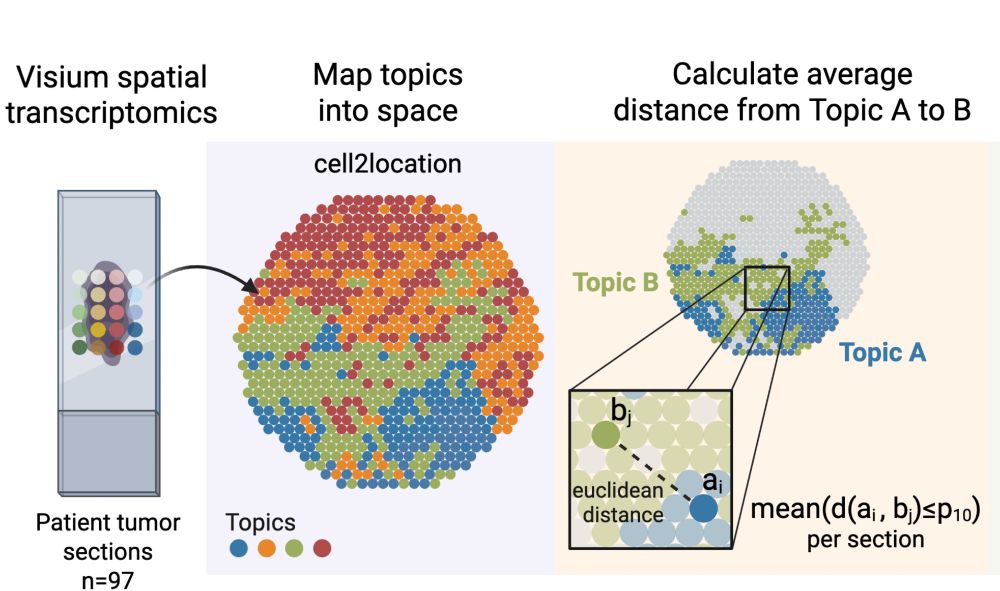
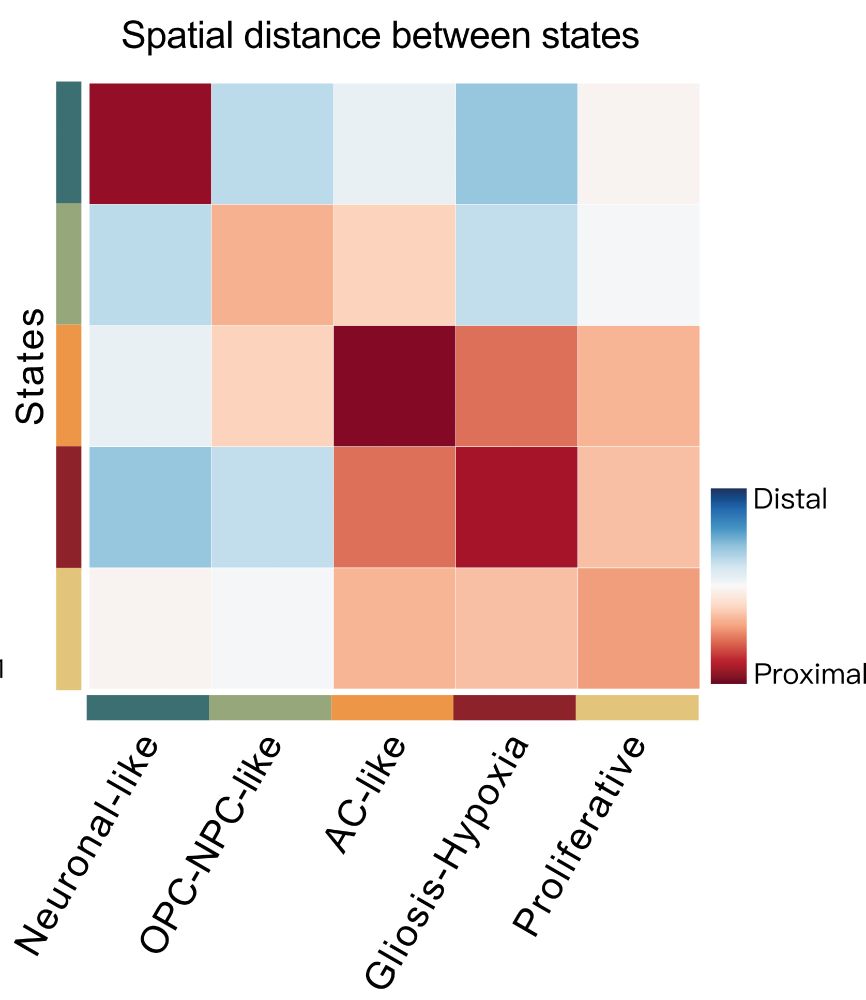
9/ Remarkably, our regulatory roadmap explains tumor architecture! States with easy transitions exist in close spatial proximity, while states separated by regulatory barriers are spatially distant. The regulatory rules we've uncovered directly shape how tumors are organized!
16.05.2025 10:04 — 👍 2 🔁 0 💬 1 📌 0
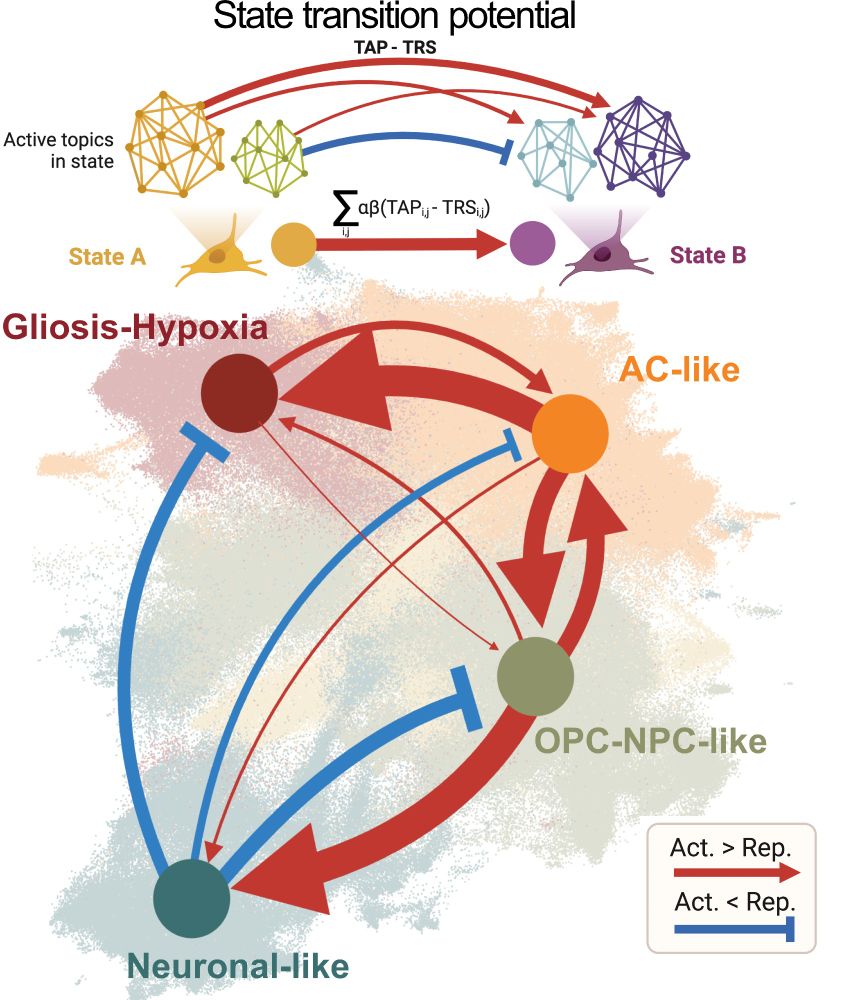
8/ This roadmap revealed striking asymmetry in GB plasticity: • OPC/NPC-like and AC-like states can easily activate multiple alternate fates • Neuronal-like states are "locked" by strong repression barriers • Transitions follow preferred directions (OPC→Neuronal easier than Neuronal→OPC)
16.05.2025 10:04 — 👍 2 🔁 0 💬 1 📌 0
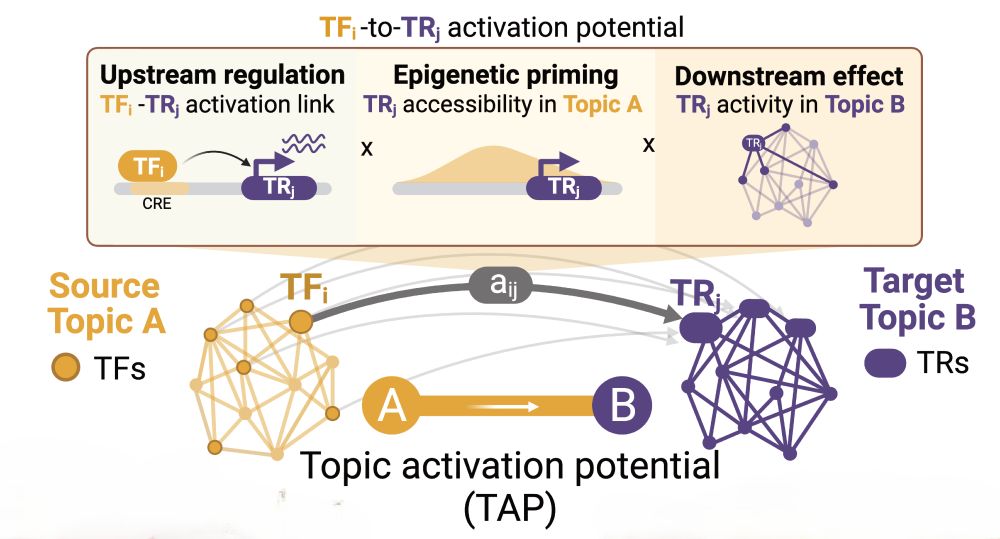
7/ 🔑 The big question: Which tumor states can easily transition to others? We developed metrics to quantify both activation potential (what enables transitions) and repression barriers (what prevents them), creating the first regulatory roadmap of GB plasticity.
16.05.2025 10:04 — 👍 2 🔁 0 💬 1 📌 0
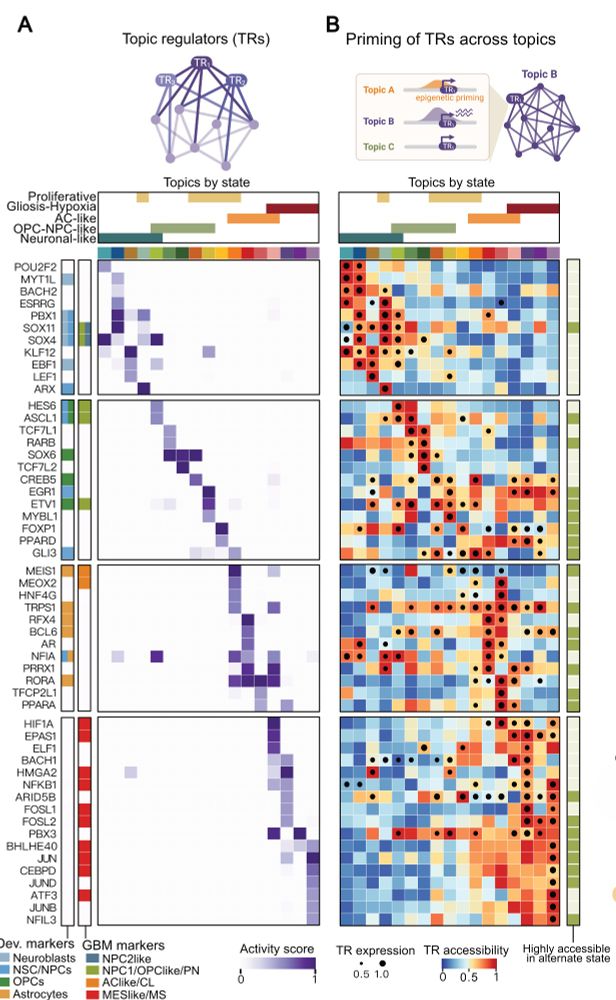
6/ The power of multi-omics: We can distinguish what a cell IS versus what it COULD BECOME. While only 16% of Topic Regulators are expressed across different tumor states, over 54% are epigenetically accessible—revealing "primed drivers" ready for activation during transitions!
16.05.2025 10:04 — 👍 2 🔁 0 💬 1 📌 0
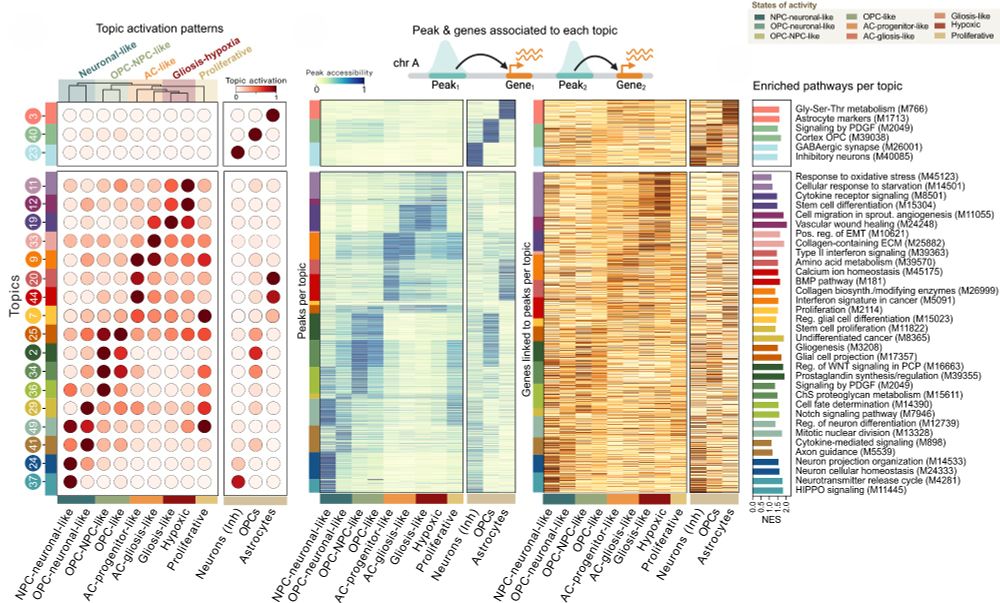
5/ Applied to GB, scDORI uncovered Topics that redefine tumor heterogeneity through regulatory logic. Each Topic links specific TFs, enhancers, and target genes that work together across tumor states. We identified key "Topic Regulators" (TRs)—master TFs for each Topic.
16.05.2025 10:04 — 👍 3 🔁 0 💬 1 📌 0
4/ scDORI: • Scales to millions of cells • Models continuous cell state GRNs • Incorporates both activation AND repression signatures • Each cell is modeled as a mixture of regulatory Topics • Can be applied to ANY multi-omic dataset (happy to hear your feedback, separate 🧵 on soon!)
16.05.2025 10:04 — 👍 3 🔁 0 💬 1 📌 0
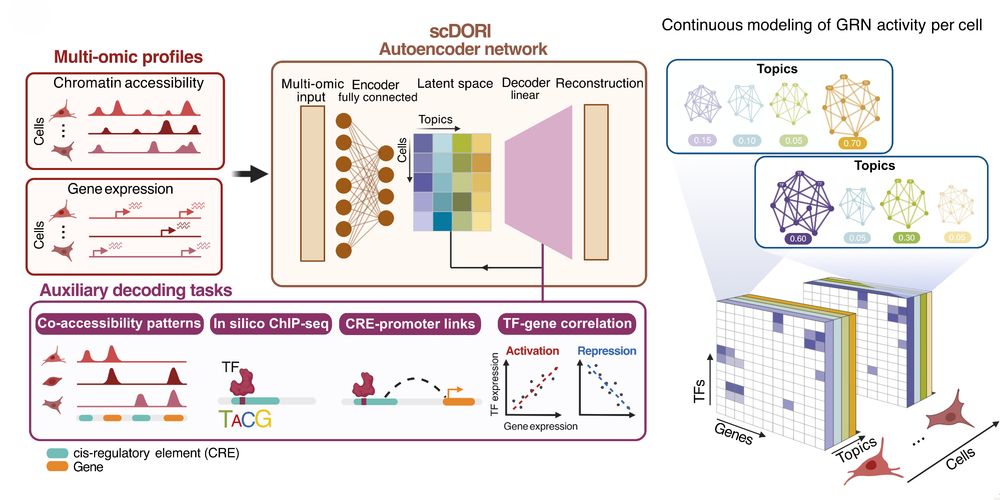
3/ To decode GB's regulatory logic, we developed scDORI—an autoencoder that decomposes multi-omic profiles into "regulatory Topics." Each Topic represents specific TF-target gene relationships, modeling cells without requiring predefined cell-types. Code: github.com/bioFAM/scDoRI
16.05.2025 10:04 — 👍 3 🔁 0 💬 1 📌 0
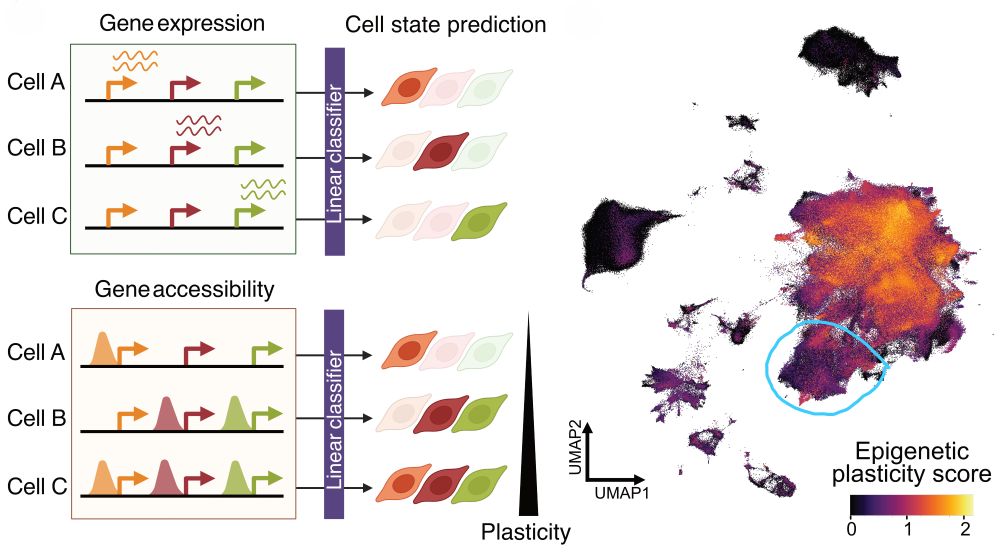
2/ Key finding: Malignant GB cells are dramatically more plastic than non-malignant cells, BUT neuronal-like tumor states show surprisingly LOW plasticity. This hints at regulatory constraints we could potentially exploit therapeutically!
16.05.2025 10:04 — 👍 3 🔁 0 💬 1 📌 0
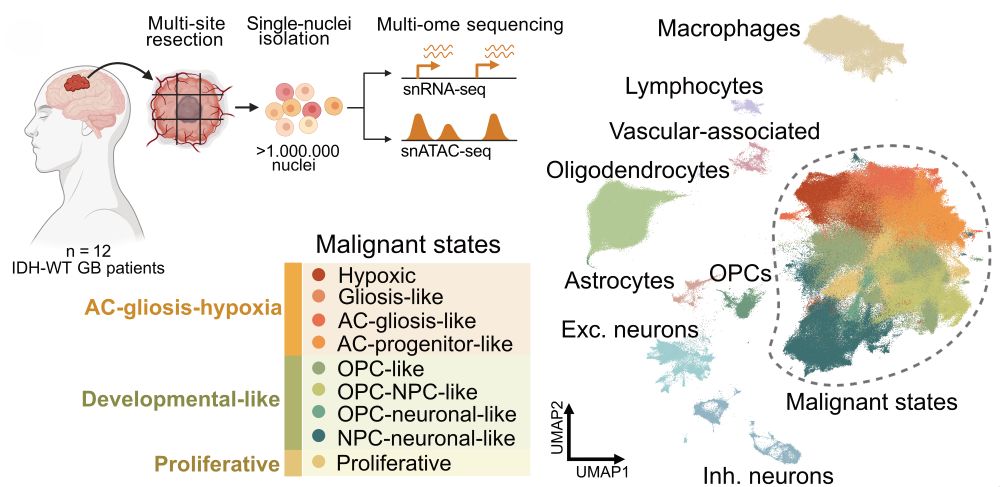
1/ Glioblastoma is driven by cellular plasticity—tumor cells switching between states. With @bayraktarlab.bsky.social, we profiled >1 M nuclei with single-cell multi-ome (RNA+ATAC) from 12 GBs, capturing the full tumor heterogeneity. We asked: what regulatory mechanisms guide these transitions?
16.05.2025 10:04 — 👍 6 🔁 0 💬 1 📌 0
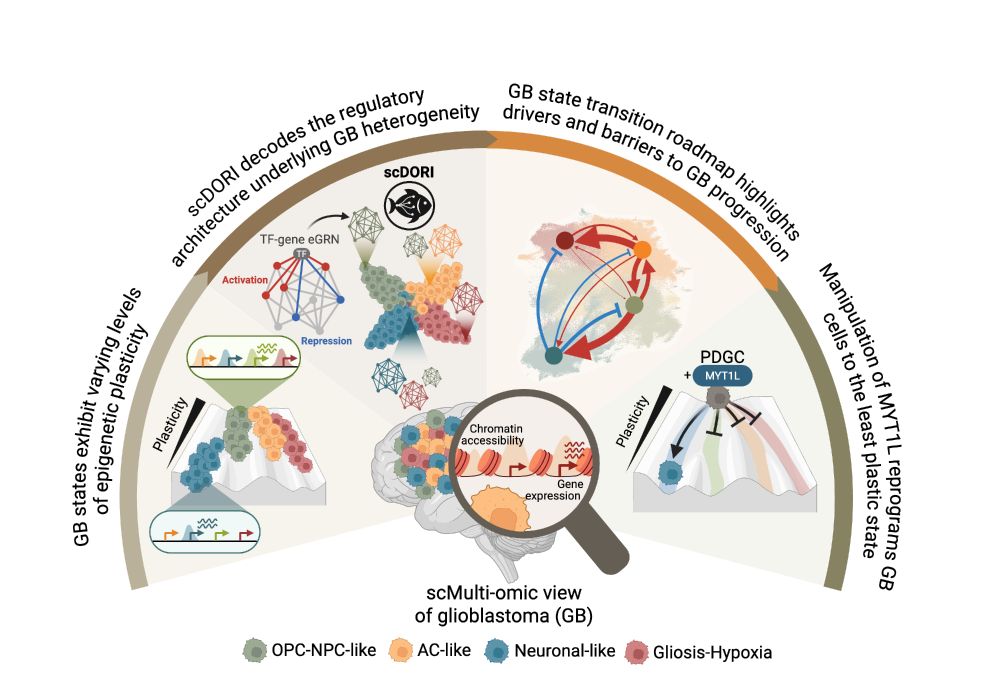
🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with @bayraktarlab.bsky.social @oliverstegle.bsky.social and @moritzmall.bsky.social, Preprint at end🧵👇
16.05.2025 10:04 — 👍 78 🔁 29 💬 1 📌 6

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
08.05.2025 16:06 — 👍 139 🔁 58 💬 4 📌 8
Genetics, bioinformatics, comp bio, statistics, data science, open source, open science!
assistant professor at ucsf interested in genetics, statistics, etc…
jeffspence.github.io
PhD student at Fraticelli lab @irbbarcelona.org
I try to convert coffee into code and ideas for uncovering biological mysteries.
Some interests: single-cell, lineage tracing, computational biology, cellular variability, premalignancy, resistance...
PhD candidate studying cohesin in context of MDS, AML.
Scientific interests :Bioinformatics, Statistics,immune-system.
Incoming Assistant Professor @ MIT Biology, Koch Institute, and Institute for Medical Engineering and Science. Single-cell advisor to Vevo Therapeutics. Lineage tracing & cancer evolution. Website: thejoneslaboratory.com
Computational genomics PhD student at Johns Hopkins BME
Images, AI, ML, and ALife.
Former: @hampshirecolg
@brandeisuniversity.bsky.social
@harvardmed.bsky.social
@uidaho.bsky.social
@hhmijanelia.bsky.social
@mdc-berlin.bsky.social
@ORNL.
Now @biohub.org.
Opinions are mine.
PhD Candidate at GeorgiaTech
Population Genetics | Cancer Evolution
The Gregory lab @DukeU studies disease mechanism using scRNAseq & spatial molecular technologies. Focus on brain tumors, Alzheimer's and MS 🧠 Home to the Molecular Genomics Core (MGC) - https://dmpi.duke.edu/cores/molecular-genomics-core-mgc
#compbio & #cancer | visuals & organisation
bioinformatician @ umm | prev phd @ dkfz
🏋🏻♀️🎨 🐈⬛ 🦅 💅🏻
Bluesky newbie. Twitter exile. Previously Professor of Diabetes in Oxford, now heading human genetics at Genentech. Living the California dream. Views my own. #YNWA
CS PhD at @columbiauniversity.bsky.social
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Statistician doing genomic data science, faculty the University of Chicago, Korean, Argentinean, American. Love kimchi, math, science, books with beautiful prose.
Group leader @crg.eu | Cell fate engineering and single-cell technology | A Taiwanese 🇹🇼
I study how genome sequence controls gene expression at Cornell.
Head of Genome Biology Dept @EMBL,
Scientist, Principal investigator, Professor
Exploring genome regulation during development,
and everything to do with enhancers.
3D genome, chromatin topology,
cell_fate, embryonic Development,
Single Cell genomics
Physicist fascinated by biology, trying to understand how cells work. Also: public engagement, science and culture, puzzles.
Associate Professor at the Biostatistics Unit of the Cyprus Institute of Neurology and Genetics (https://www.cing.ac.cy/en/about-us/biostatistics_unit)