Ohh curious to see your work too then :)
10.06.2025 08:18 — 👍 0 🔁 0 💬 0 📌 0

GitHub - theislab/mapqc: MapQC - a metric for the evaluation of single-cell query-to-reference mappings
MapQC - a metric for the evaluation of single-cell query-to-reference mappings - theislab/mapqc
MapQC is a pip-installable python package, and runs in less than 2 mins on a query dataset of 30k and a ref. of 0.5M cells. For more info, see our GitHub repo github.com/theislab/mapqc. It also includes tutorials. Try it out, and let me know what you think!
03.06.2025 08:24 — 👍 0 🔁 0 💬 0 📌 0

Re-mapping with different parameters drastically improves the mapping, and enables identifying disease-specific cell states, such as altered smooth muscle cells in lungs of patients with IPF (reproducible across studies). MapQC thus helps you make the best use of your data!
03.06.2025 08:24 — 👍 2 🔁 0 💬 1 📌 0

Here’s an example of a mapping that failed. For some cell types (e.g. circled ct), the UMAP suggests the query mixes well with the reference. However, mapQC scores show this is not the case, and downstream-analysis indeed results in batch-effect driven conclusions.
03.06.2025 08:24 — 👍 0 🔁 0 💬 1 📌 0

This results in cell-level mapQC scores, with a score >2 indicating large distance to the reference. We expect controls in the query to be the same as in the ref., i.e. to show scores <2. In contrast, disease samples should show some high distance to the ref. (local scores >2).
03.06.2025 08:24 — 👍 0 🔁 0 💬 1 📌 0

How does it work? We use the control samples in the large-scale reference to obtain prior knowledge of normal inter-sample variation. We do this locally, such that we learn cell-state-specific inter-sample distances. We compare those to query sample distances to the reference.
03.06.2025 08:24 — 👍 1 🔁 0 💬 1 📌 0

We therefore developed mapQC, a method that takes as its input any query mapping to a large-scale reference, and outputs a cell-level mapQC score. The score will tell you if, and where, the mapped query contains batch effects, or if e.g. disease-specific variation was removed.
03.06.2025 08:24 — 👍 0 🔁 0 💬 1 📌 0

One commonly used metric, LISI, is highly sensitive to cell numbers, which in fact are independent of integration/mapping quality and should not affect metric outcome. Finally, all of the existing metrics lack a clear rationale for a cutoff between good and bad mappings.
03.06.2025 08:24 — 👍 0 🔁 0 💬 1 📌 0

Moreover, standard integration and mapping metrics fail to pinpoint these failures: they quantify the wrong things. Here’s an example, with one very poor and one good-quality mapping resulting in the same scores for several metrics, but not mapQC.
03.06.2025 08:24 — 👍 1 🔁 0 💬 1 📌 0
With the surge in large-scale single-cell atlases, many people have started using atlases to analyze their new data. However, query-to-reference mappings, used to combine a reference with new data, often do not produce a good embedding. This leads to data misinterpretation.
03.06.2025 08:24 — 👍 0 🔁 0 💬 1 📌 0
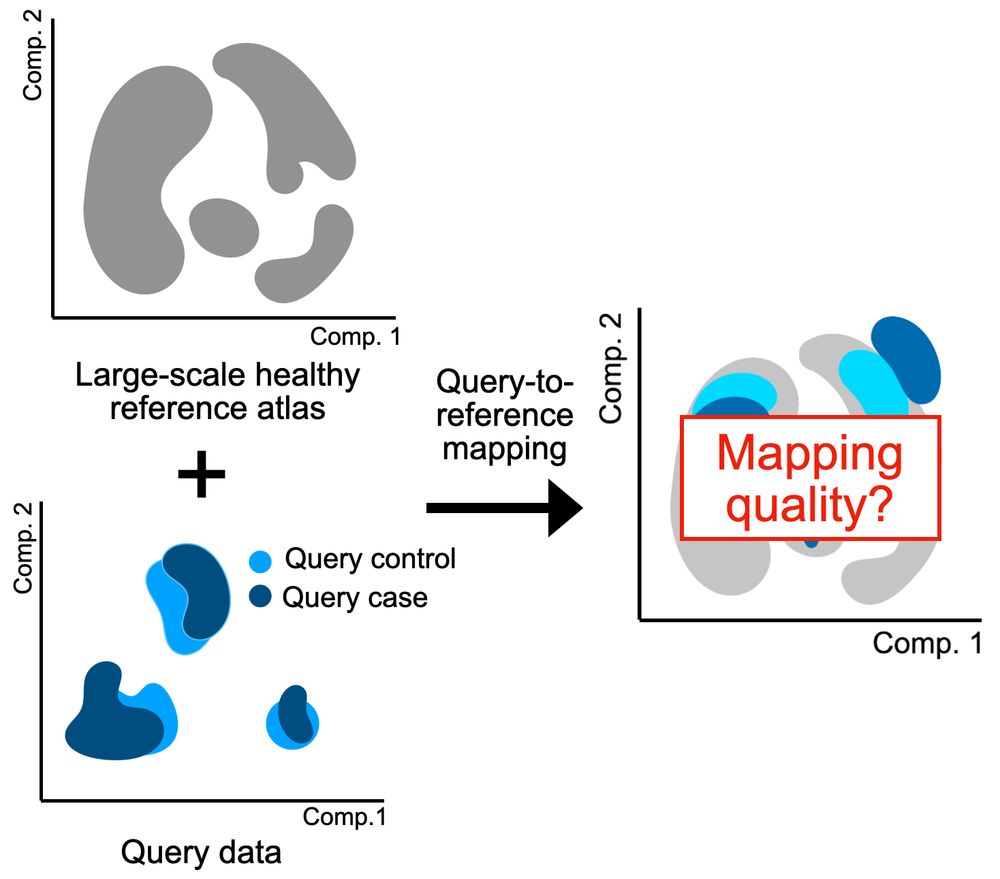
Analyzing your single-cell data by mapping to a reference atlas? Then how do you know the mapping actually worked, and you’re not analyzing mapping-induced artifacts? We developed mapQC, a mapping evaluation tool www.biorxiv.org/content/10.1... from the @fabiantheis lab. Let’s dive in🧵
03.06.2025 08:24 — 👍 24 🔁 10 💬 2 📌 0
7/7 We hope that our guide will help you to construct high-quality atlases and efficiently explore their contents. We would love to hear your thoughts on atlasing and your comments on our guide!
13.12.2024 10:32 — 👍 1 🔁 0 💬 1 📌 0
6/7 When building atlases, the downstream use-cases should always be the primary focus. Thus, we extensively discuss how atlases can be used in single-cell and broader biological research.
13.12.2024 10:32 — 👍 1 🔁 0 💬 1 📌 0
5/7 Atlas building involves many steps: defining the atlas’ focus, data preprocessing, integration, annotation and evaluation. Afterwards the atlas must be shared with the community and eventually updated and extended. We present diverse considerations associated with each step.
13.12.2024 10:32 — 👍 1 🔁 0 💬 1 📌 0
4/7 However, constructing an atlas is not straightforward and the methods in the field are still evolving. With this in mind, we discuss different approaches for atlas building, along with their pros and cons. This will help you make better informed decisions in upcoming atlasing projects.
13.12.2024 10:32 — 👍 1 🔁 0 💬 1 📌 0

3/7 Atlases have value beyond individual datasets. Their diversity in samples, donors and conditions paint a more holistic picture of biology. Moreover, they are invaluable as a reference for analyzing new data, easing preprocessing and guiding interpretation.
13.12.2024 10:32 — 👍 2 🔁 0 💬 1 📌 0
2/7 The surge in single-cell datasets improved our understanding of biology, and integrating these datasets into unified “atlases” can teach us even more: we can create consensus cell type naming, increase power for learning disease-related patterns, and compare across multiple diseases.
13.12.2024 10:32 — 👍 1 🔁 0 💬 1 📌 0

1/7 Planning to build a single-cell atlas? Or wondering how atlases can be useful to your research? Read our guide on single-cell atlases www.nature.com/articles/s41... published in Nature Methods, by @lisasikkema.bsky.social, @khrovatin.bsky.social, Malte Luecken, @fabiantheis.bsky.social et al.
13.12.2024 10:32 — 👍 50 🔁 17 💬 1 📌 3
Wellcome Trust Clinical PhD Fellow in the Garnett Lab @sangerinstitute.bsky.social. Interested in cancer plasticity, multiplexed assays, ML, & comp bio.
https://ttszen.com/
Scientist at Max Planck Genome Centre. Genomics/Single cell sequencing/Flow Cytomery&Cell Sorting. Microscale farmer and owner of TheMostSpoiledCatinTheWorld.
Computational Biologist studying pancreas using single cell genomics.
Senior Scientist @ The University of Sydney: cytometry, CyTOF, single-cell & spatial multiomics, systems immunology. Interests in respiratory 🫁 & neurological 🧠 infectious disease 🦠, paediatric immunology 👶.
https://tomashhurst.github.io
Professor in Systems Biology & Genetics @EPFL, opinions my own; Single Cell Omics / Gene Regulation / Transcription Factor / Stem Cells / Regulatory Variation / ML / Imaging / Adipose Biology / Microfluidics
https://www.epfl.ch/labs/deplanckelab
Bioinformatician, data scientist, software developer
Also @_lazappi_ and @lazappi@mastodon.au
Co-director of MoPED (Mechanisms of Paracrine & Endocrine Disorders) lab in Marseille 🇫🇷 on cell signaling & determination of self & beyond during neural crest differentiation. Personal account.
Computational biologist in Raphael Gottardo's group at the Lausanne University Hospital (CHUV)
Dad, immunologist, and aikidoka. Scientific director at Immune Health https://www.med.upenn.edu/i3h/
-Personal account-
Bioinformatics PhD, cheminformatics, data scientist, statistics, toxicology and coding at Eli Lilly... I do it all! Currently focused in off-target ID for genetic medicines. Mom to a kid and a cat. Thoughts and opinions are my own
data science postdoc in Tübingen 🧬🖥️🧠 scRNA data analysis, UMAP/tSNE & retina neuroscience | science journalism on AI & sustainability 🤖❤️🌍 | easily sidetracked by small plot details & cool birds 📈🔍🦜
‖ Permanent Researcher / INRIA Start. Faculty @ INRIA Rennes 🇫🇷 ‖
BioInfo/CompBio: algorithms, genomics, pathogens & rapid diagnostic of antibiotic resistance《 https://brinda.eu | https://github.com/karel-brinda 》
PhD student Genetics+CS @Stanford w/ @anshulkundaje.bsky.social | Prev CS @telecomparis @Columbia @Microsoft | I'm interested in using AI/ML to model biological systems 👩🏻💻🧬☕ | valehamiri.com
MD student with Herbert Schiller @www.helmholtz-munich.de | MSc Bioinformatics @edinburgh-uni.bsky.social | aspiring physician scientist - interested in PCCM, lung fibrosis, and single cell omics | mountaineering enthusiast | Views are my own
Postdoc between Helmholtz Munich and ETH Zurich | Theis Lab & Treutlein Lab | Computational Biology, Machine Learning & Organoids
PhD student at Merad lab @Mount Sinai | prev MSc at Sallusto lab, Becher lab @ETH Zürich & Apotheker @FU Berlin
Bioinformatics, gene expression, software development, QTL causal gene prioritization. Wageningen University.
PhD Student in Computational Biology at TU Munich (Gagneur lab) and Helmholtz Munich (Theis lab)
Interested in rare variants and their effect in Population-scale cohorts










