Happy to see this work out now in Genome Biology! Check out the final version here for your FACS DMS needs: link.springer.com/article/10.1...
26.02.2026 23:21 — 👍 11 🔁 5 💬 0 📌 1Happy to see this work out now in Genome Biology! Check out the final version here for your FACS DMS needs: link.springer.com/article/10.1...
26.02.2026 23:21 — 👍 11 🔁 5 💬 0 📌 1
A wonderful first for me at the upcoming @biophysicalsoc.bsky.social meeting in SF is having many lab members present! See 6 brilliant graduate students postdocs from the lab present talks and posters on how they are pushing the boundaries of technology and mechanistic membrane protein biology
20.02.2026 18:26 — 👍 15 🔁 4 💬 1 📌 0Congratulations!! 🥳
21.07.2025 21:37 — 👍 1 🔁 0 💬 0 📌 0Check out Jerome's paper! As we push the boundaries of DMS, it is critical that we develop robust ways to score variant effects--and he is doing just that. Excited to put this to use in our upcoming GPCR-DMS endeavors :)
30.06.2025 16:18 — 👍 7 🔁 1 💬 0 📌 0
We are hiring a #postdoc in the Deshpande lab! Come join us tackle important mechanistic questions in inflammatory signaling. Please share or apply!
careers.gene.com/us/en/job/20...

Beautiful work from our colleagues in the Jura & Verba labs here @ UCSF! Cryo-EM structures of PI3Ka / KRas complex in context of lipid nanodiscs. Read on for unexpected dimers & to see how high-rez structural biology w/ the right reconstitutions can inform mechanism! www.biorxiv.org/content/10.1...
26.03.2025 16:20 — 👍 12 🔁 4 💬 0 📌 0
It is important at this pivotal moment to @standupforscience.bsky.social. Looking forward to sharing my thoughts on why this should be a national priority at tomorrow’s rally in SF:
05.03.2025 16:54 — 👍 275 🔁 89 💬 2 📌 4
New preprint! Here, we explore how endocytosis encodes receptor-specific cAMP/PKA signaling downstream of three endogenously coexpressed GPCRs. www.biorxiv.org/content/10.1...
27.02.2025 18:14 — 👍 8 🔁 3 💬 0 📌 0
My lab at
@stanfordmedicine.bsky.social
is recruiting! We are looking for a postdoc at the interface of quantitative proteomics and G protein-coupled receptor (GPCR) biology: postdocs.stanford.edu/prospective/...
#TeamMassSpec #Proteomics #GPCR #Postdoc
Thanks Reid! Looks like they are queued to release with the next PDB update on Jan. 15
08.01.2025 20:32 — 👍 0 🔁 0 💬 1 📌 0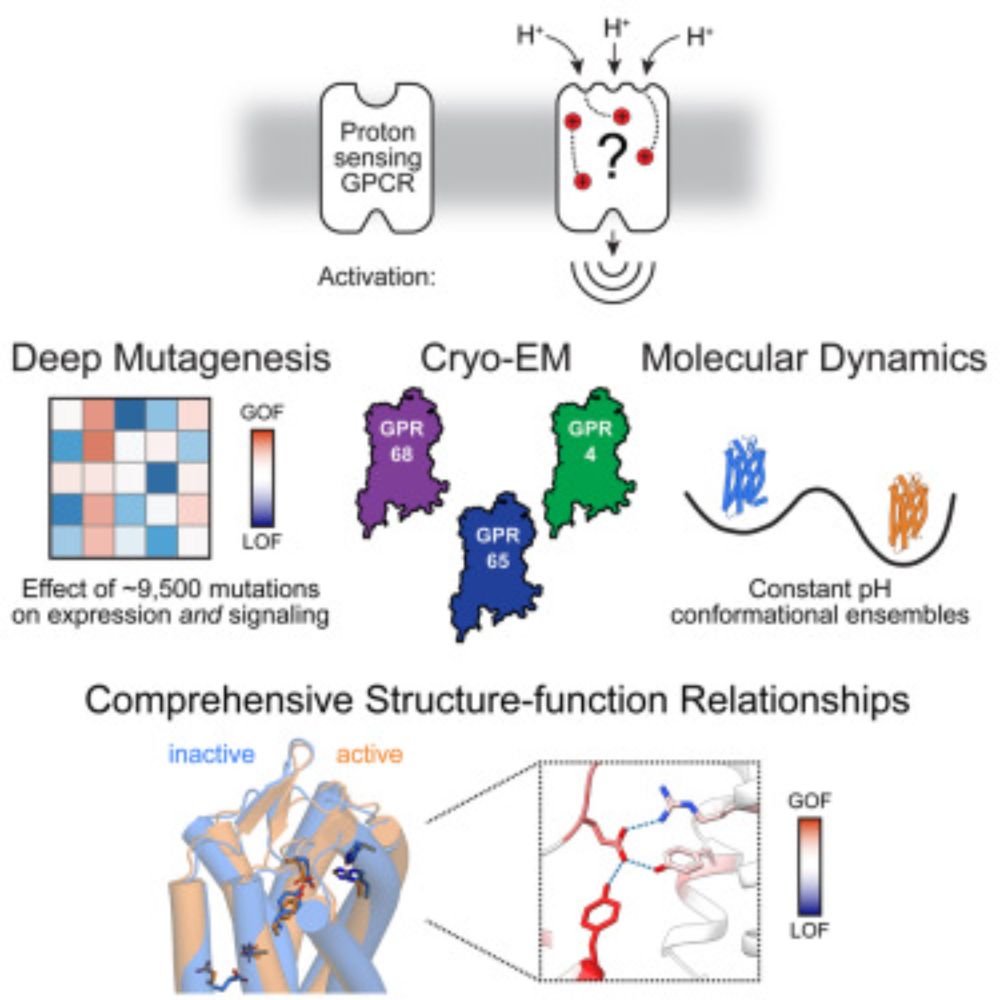
Congrats @willowcoyote.bsky.social @amanglik.bsky.social and all authors on a tour de force dissection of proton-sensing by GPCRs! DMS, cryo-EM, MD -- it has it all! www.cell.com/cell/fulltex...
02.01.2025 17:31 — 👍 32 🔁 16 💬 0 📌 0Thanks Stephanie! Hope all is going will w/your new lab - excited to see all the amazing work to come!
07.01.2025 06:16 — 👍 1 🔁 0 💬 0 📌 0Thanks Joe!
07.01.2025 06:13 — 👍 0 🔁 0 💬 0 📌 0Thanks, glad you enjoyed it!
07.01.2025 06:08 — 👍 1 🔁 0 💬 0 📌 0Thanks Alex!
07.01.2025 06:06 — 👍 0 🔁 0 💬 0 📌 0Thanks for the invite! Excited to share some of our recent work tomorrow 😀
06.01.2025 20:35 — 👍 2 🔁 0 💬 0 📌 0Tagging all the folks who I can find here... @amanglik.bsky.social @willowcoyote.bsky.social @justingenglish.bsky.social @delemottelab.bsky.social ...
06.01.2025 20:27 — 👍 0 🔁 0 💬 0 📌 0Thanks @justingenglish.bsky.social!! Awesome working with you and excited to see where we go in the future 👀
06.01.2025 20:01 — 👍 2 🔁 0 💬 0 📌 0There is certainly more than can be shared here, but I will end it there with a HUGE thanks to everyone involved! This was both a challenging and fun project to be involved in with many great folks without which none of this would be possible. (13/13)
06.01.2025 19:57 — 👍 2 🔁 0 💬 3 📌 0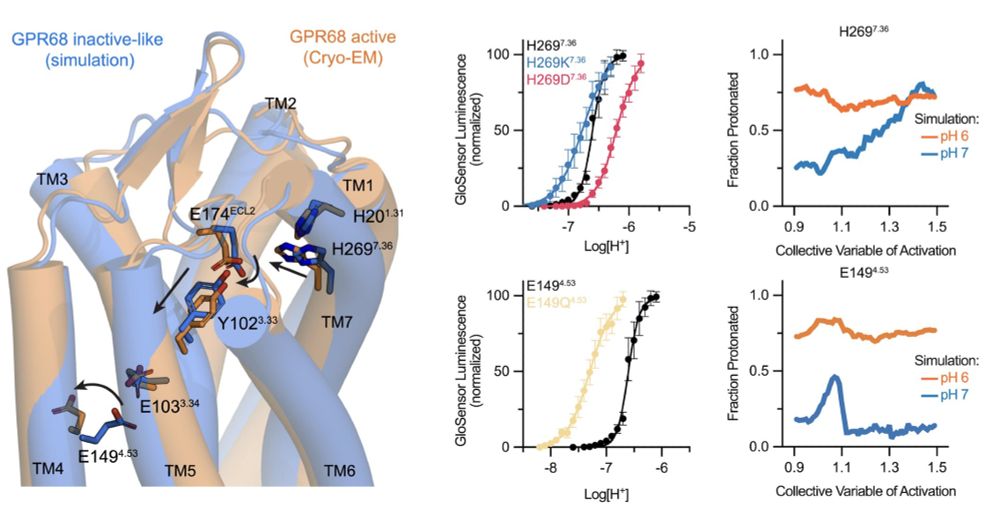
Ultimately, this enabled us to identify residues which change pronation with conformation and pH. Combining our DMS, cryo-EM, MD, and extensive pharmacologic validation (by XP Huang), we were able to devise a comprehensive model for pH activation of GPR68 (12/n)
06.01.2025 19:57 — 👍 2 🔁 0 💬 1 📌 0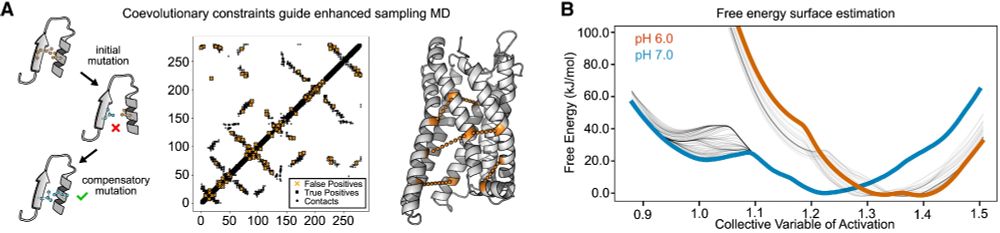
Well, Darko went above and beyond. He developed a new method which uses coevolutionary information to identify both alternative receptor conformations AND the effect of pH on the conformational ensemble. (11/n)
06.01.2025 19:57 — 👍 3 🔁 0 💬 1 📌 0
We needed an inactive-state structure model to fully interpret our results, so we teamed up with
Darko Mitrovic (in @delemottelab.bsky.social lab) to do some molecular dynamics simulations of GPR68 (10/n)
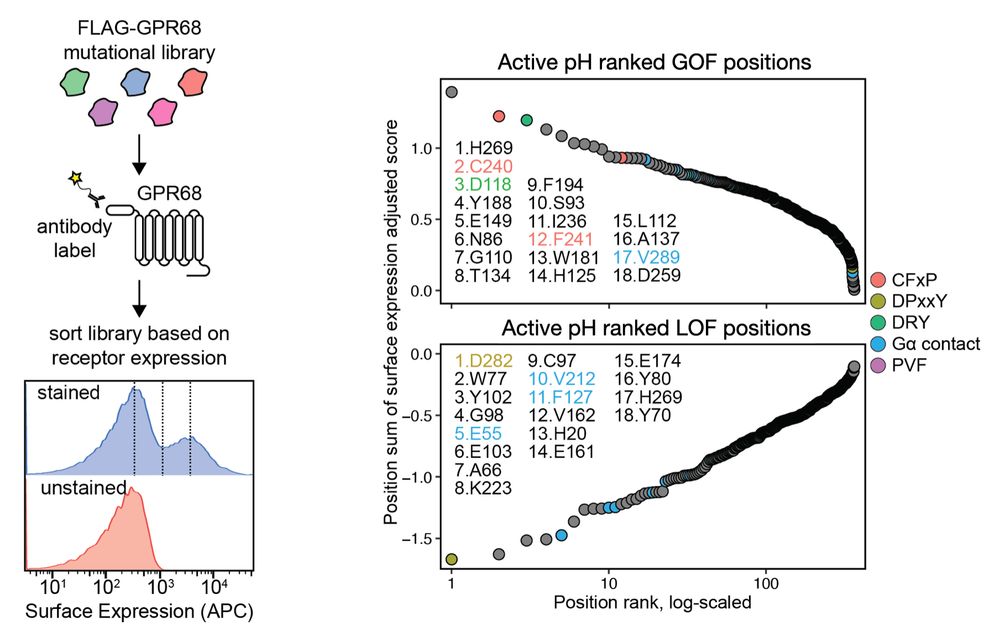
We followed up with a surface expression screen to determine the which mutations alter receptor expression. This allowed to deconvolve the mutational effects on expression vs activation to uncover GOF and LOF activity specific to activation. (9/n)
06.01.2025 19:57 — 👍 1 🔁 0 💬 1 📌 0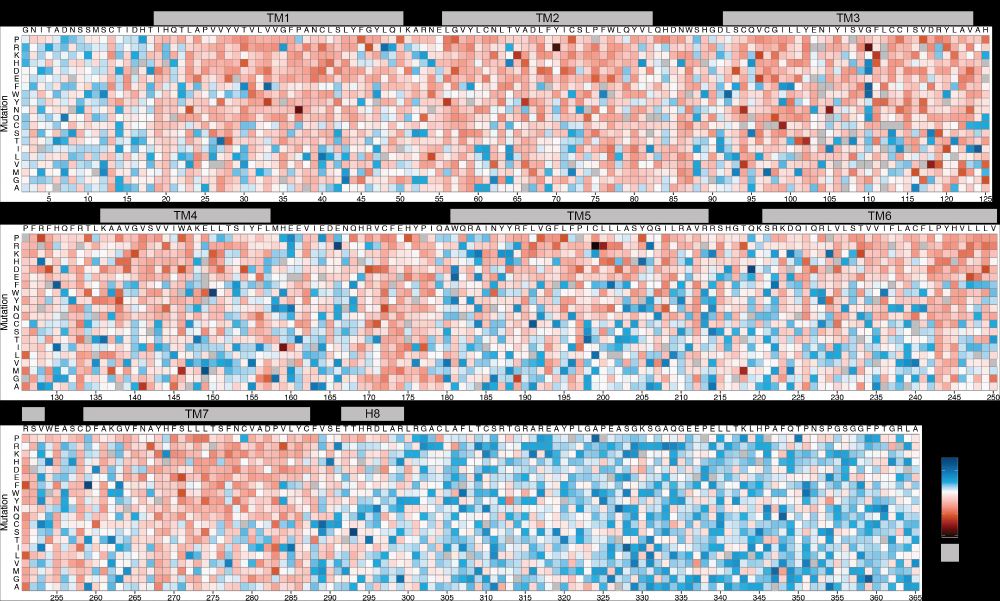
The result: a comprehensive map of each mutation's effect on pH activation. (8/n)
06.01.2025 19:57 — 👍 2 🔁 0 💬 1 📌 0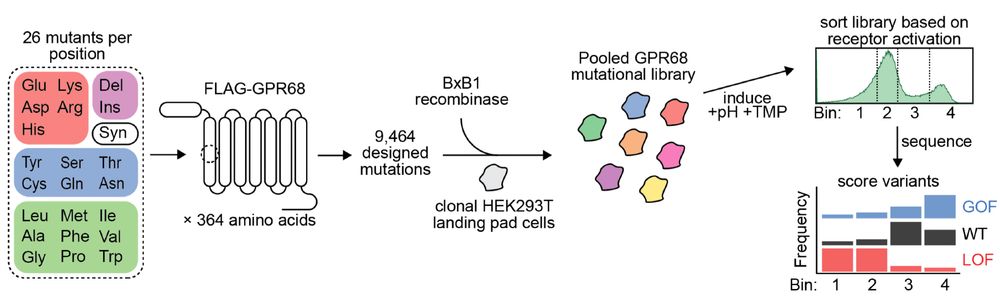
We then generated a mutational library of GPR68 using the DIMPLE platform developed in @willowcoyote.bsky.social's lab and screened it using our cAMP FACS assay (7/n)
06.01.2025 19:57 — 👍 1 🔁 0 💬 1 📌 0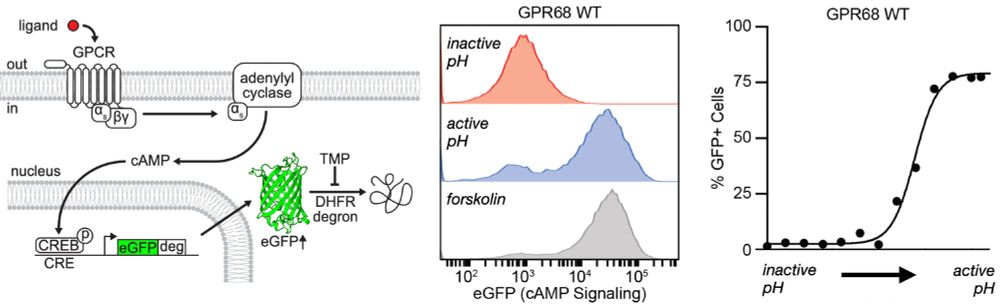
Using GPR68 as our model, we set out to use mutational scanning to determine the effect of every mutation on proton activation. We developed a new FACS-based method to measure Gs coupled receptor activation (enabled by a new TRE's from @justingenglish.bsky.social ) (6/n)
06.01.2025 19:57 — 👍 2 🔁 0 💬 1 📌 0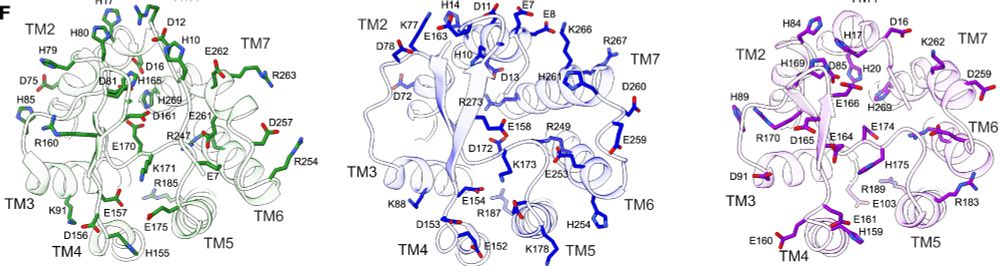
We needed to ascribe a functional role for each residue in proton activation. Unfortunately, these receptors are littered with an enormous number polar and charged residues which may be implicated. (5/n)
06.01.2025 19:57 — 👍 1 🔁 0 💬 1 📌 0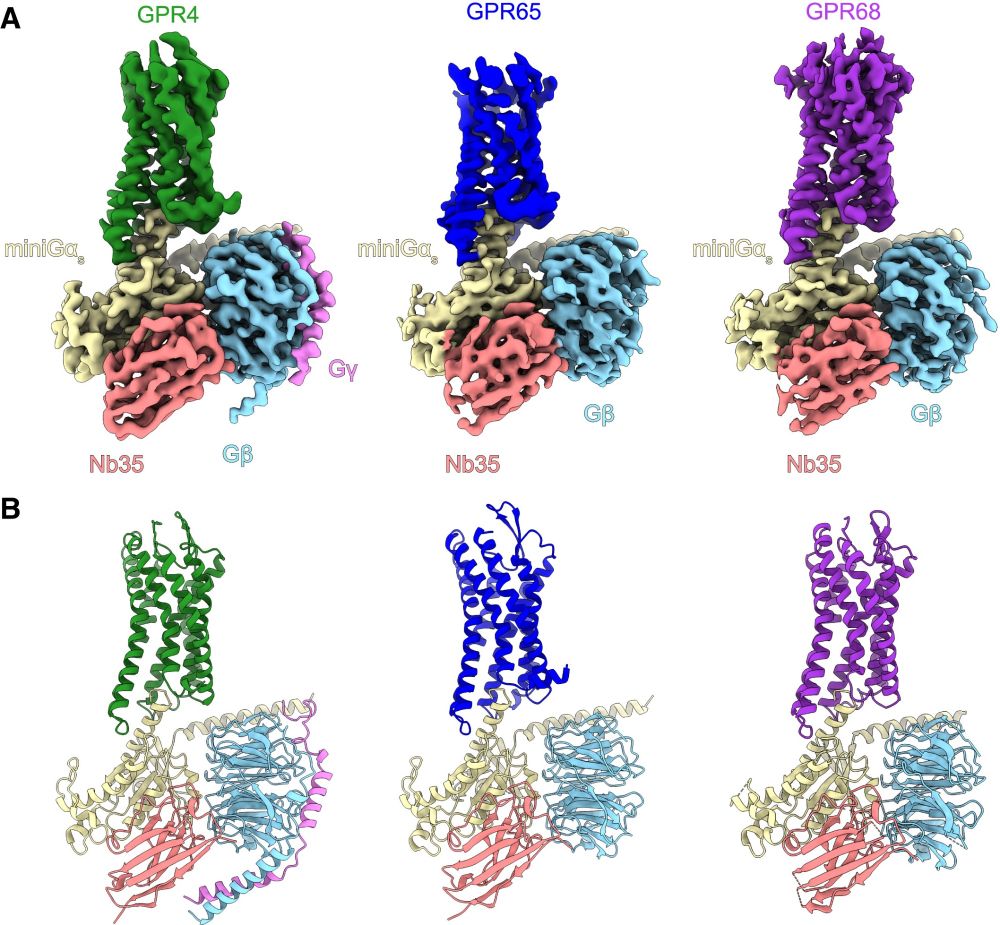
To start, Nick Hoppe (in @amanglik.bsky.social's lab) set out to determine the 3D architecture of each human proton sensor (GPR4, GPR65, and GPR68) using cryo-EM. These provided insights into the architecture and arrangement of putative proton-sensing residues in the active-state (4/n)
06.01.2025 19:57 — 👍 1 🔁 0 💬 1 📌 0We wanted to determine the location and identities of all residues involved in coordinating protons to drive receptor activation. (3/n)
06.01.2025 19:57 — 👍 1 🔁 0 💬 1 📌 0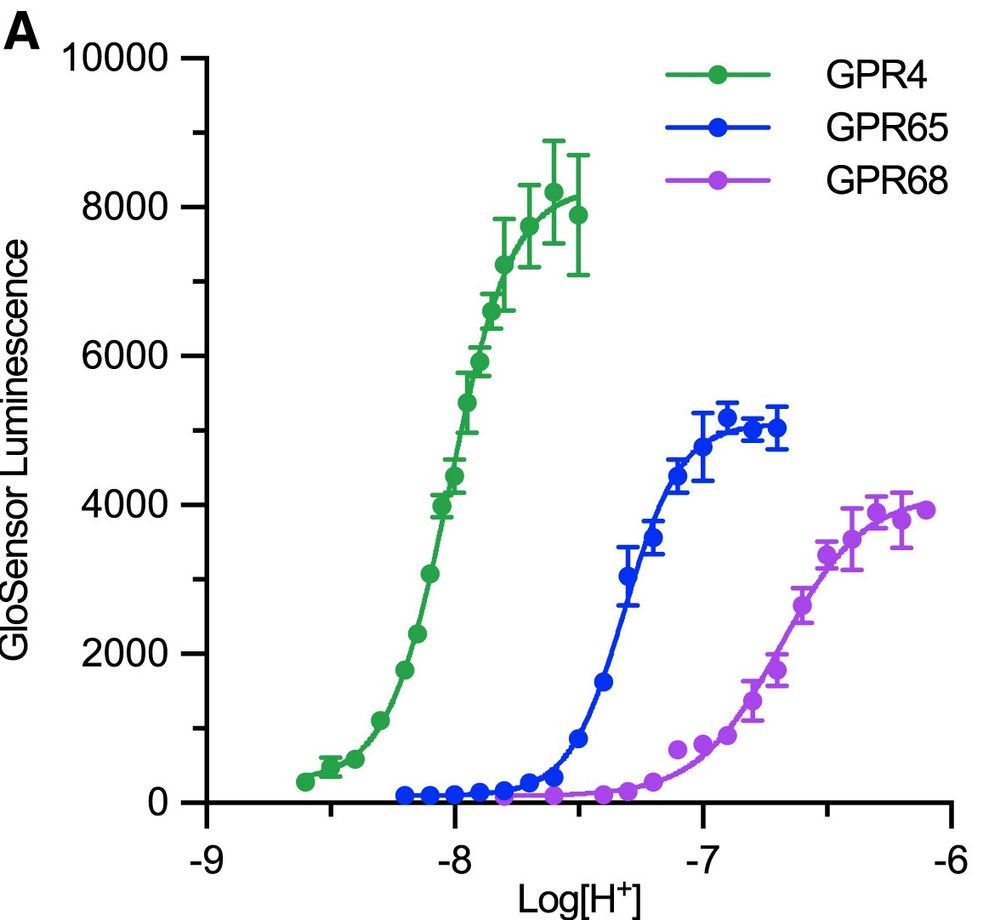
We have known for some time that several GPCRs respond to changes in pH. However, unlike small molecules, peptides, and other stimuli, protons are a bit wonky--we can't directly see them with standard approaches. (2/n)
06.01.2025 19:57 — 👍 2 🔁 0 💬 1 📌 0