It's hard to express how much gratitude I have to everyone involved in the journey, both mine and @napari.org's, that has led to this point. #EurekaPrizes A small attempt below: 👇
01.08.2025 07:29 — 👍 29 🔁 14 💬 4 📌 1
napari roadmap blog post — napari blog documentation
🚀The napari team is excited to announce our brand new development roadmap — a community-informed vision for the future of napari!
🔍Read the blog to learn about our process and priorities:
napari.org/island-dispa...
🗺️OR check out the roadmap itself on napari.org:
napari.org/stable/roadm...
10.07.2025 20:03 — 👍 23 🔁 5 💬 0 📌 1
If you post the frames in a thread then we can try scrolling really fast
02.07.2025 12:47 — 👍 1 🔁 0 💬 0 📌 0
bsky.app/profile/wais...
10.06.2025 13:51 — 👍 0 🔁 0 💬 1 📌 0
There is a @napari.org hackathon at GloBIAS!
Be there or be square (or whatever layer type you'd like)
16.05.2025 11:24 — 👍 2 🔁 1 💬 0 📌 0
TFW you discover the internet in the conference venue isn't stable enough for a Google Slides presentation with videos 😅 luckily with >24hr warning
06.05.2025 12:15 — 👍 3 🔁 0 💬 1 📌 0
Python Segmentation API — polarityjam documentation
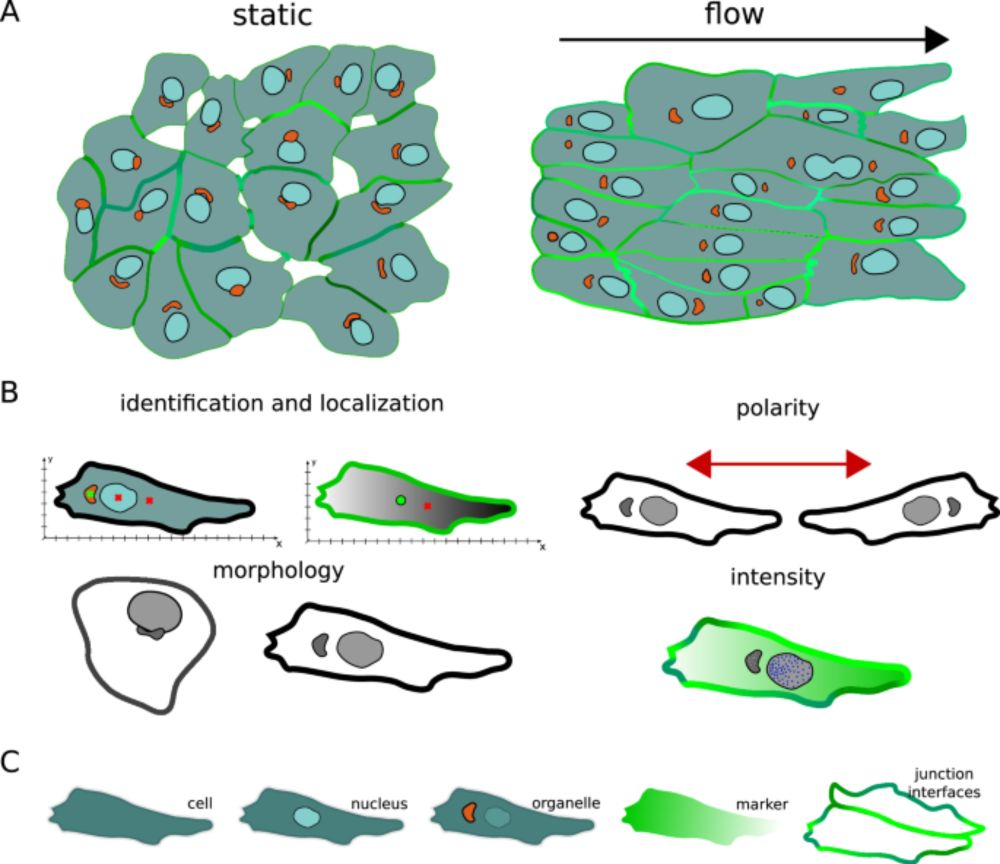
We have a suite for studying cell polarity in Python polarityjam.readthedocs.io feel free to post an issue if you need any more functionality
26.04.2025 12:27 — 👍 2 🔁 0 💬 1 📌 0
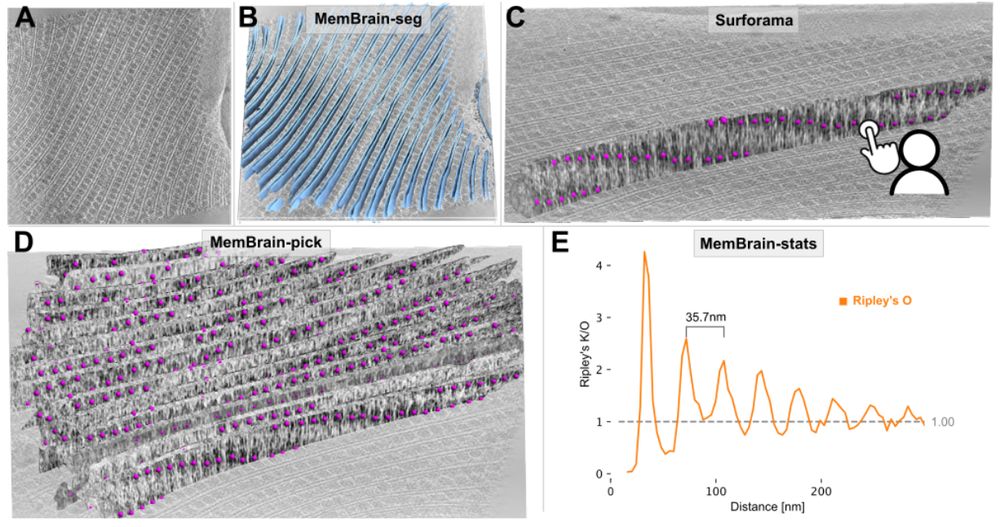
Figure 4. MemBrain v2 end-to-end workflow detects periodic phycobilisome organization. A: Raw tomogram slice of EMD-31244. B: Out-of-the-box MemBrain-seg segmentation (light blue). C: A single membrane instance can be visualized in Surforama and manually annotated with GT phycobilisome positions (magenta). D: MemBrain-pick localizes particles (trained with data from C) on all membranes in the tomogram. E: MemBrain-stats computes Ripley’s O statistic using the positions from D with a bin size of 5nm. The distance between peaks (35 nm) was measured to estimate chain unit spacings.
We have updated our #MemBrain v2 preprint with a lot more details about the MemBrain-pick and MemBrain-stats modules, as well as some application examples!
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...
23.04.2025 08:47 — 👍 71 🔁 20 💬 3 📌 1
Yo #TeamTomo, check out our updated #MemBrain v2 preprint. And better yet, give it a whirl on your #CryoET membranes! Please send us your feedback! 🧪🧶🧬🔬
24.04.2025 21:35 — 👍 24 🔁 3 💬 0 📌 0
"no no, I said turn up the economy"
07.04.2025 20:52 — 👍 0 🔁 0 💬 0 📌 0
Check this out!!! The results from our CryoET particle picking competition are available, the full test data (public and private datasets) is available, and you can dive right into the data on the CZ CryoET Data Portal! Enjoy and please let us know what you think :)
06.03.2025 17:18 — 👍 8 🔁 1 💬 0 📌 0
Awesome! So glad to see my former RL professor Andy Barto get this recognition (and Rich is too!)
05.03.2025 12:42 — 👍 0 🔁 0 💬 0 📌 0
Last night we were doing a family dance party...
My smartwatch gave a notification...
You've been running for 5 minutes...
I feel judged.
12.02.2025 14:18 — 👍 2 🔁 0 💬 0 📌 0
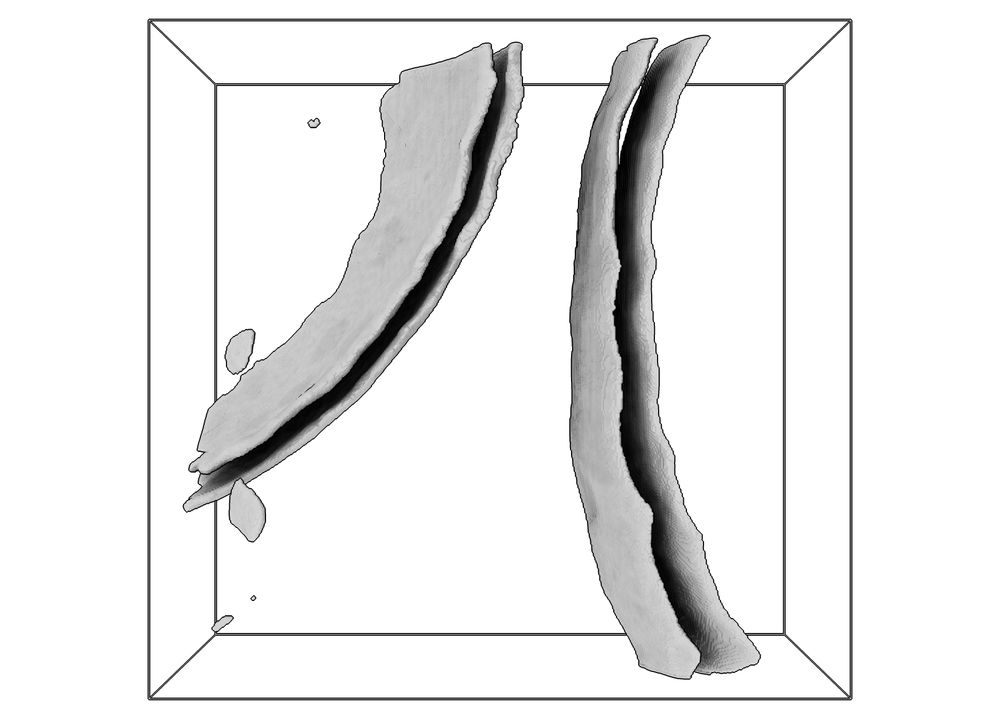
Hey #teamtomo, if you often find yourself using the excellent membrain-seg from @lorenzlamm.bsky.social et al., you might find my napari plugin "napari-segselect" useful. Let's say your tomogram contains the edges of two bacterial cells, each with a membrane and cell wall:
06.02.2025 11:02 — 👍 37 🔁 12 💬 3 📌 1
I just want to take the opportunity to remind everyone that the Hamster Dance still exists. You can even find 10 hour long videos of it. YW
05.02.2025 13:25 — 👍 0 🔁 0 💬 0 📌 0

Screencap of Drosophila courtship, caption "The male is flapping his wings"

Screencap showing Drosophila flight arena
There's a Korean soap opera on Netflix (When the Stars Gossip) which has *Drosophila courtship in space* as a major plot point (Ep 2). They have Michael Reiser & lab's flight arenas (you can see Janelia & MIchael's initials on the panels). Great way to learn about Drosophila courtship!
05.02.2025 01:51 — 👍 8 🔁 4 💬 1 📌 0
TL;DR installers are really challenging to make. The team made some tooling for @napari.fosstodon.org.ap.brid.gy that was so fantastic that it is spreading across the scientific python ecosystem!
05.02.2025 01:24 — 👍 9 🔁 3 💬 0 📌 0

Senior Research Scientist, AI x Imaging, Science
Redwood City, CA (Open to Flex)
🚨Looking for a new opportunity at the intersection of #AI and #bioimaging? Want to work with an awesome team?
We have multiple open research scientist positions @cziscience.bsky.social. Please RT!
boards.greenhouse.io/chanzuckerbe...
boards.greenhouse.io/chanzuckerbe...
25.11.2024 14:04 — 👍 2 🔁 1 💬 0 📌 0
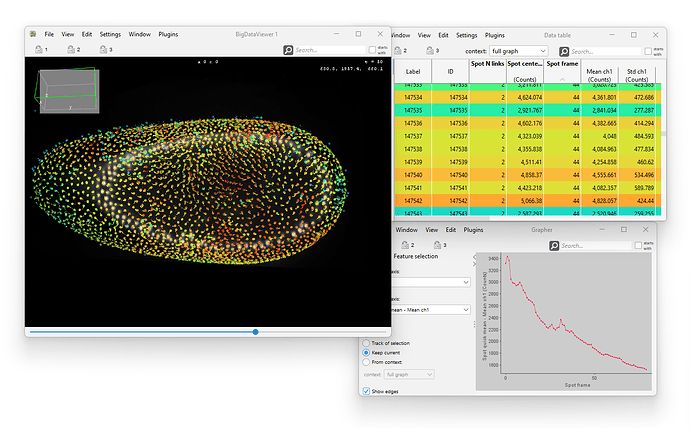
We just released and uploaded a new version of *Mastodon*, the large-scale tracking and track-editing framework for large, multi-view images.
Mastodon is available via Fiji, via a dedicated update site.
mastodon.readthedocs.io/en/latest/do...
Below is a thread that survey some of the novelties ⏬️
30.01.2025 16:44 — 👍 127 🔁 44 💬 3 📌 3
It was the user's fault for not signing up sooner.
30.01.2025 14:51 — 👍 1 🔁 0 💬 0 📌 0
🤣
30.01.2025 14:46 — 👍 0 🔁 0 💬 1 📌 0
is the strategy for getting gemini users to have pop up windows in google apps?
30.01.2025 12:56 — 👍 0 🔁 0 💬 1 📌 0
AI startup recruiting emails these days:
"Our company coming out of stealth mode w new capital. We need a founding engineer for our startup that is focused on developing foundation models of founding engineers. Our plan for monetization is replacing founding engineers in post-stealth mode companies"
23.01.2025 14:51 — 👍 5 🔁 1 💬 0 📌 0
For some reason it feels very judgy when my smart watch says "Good job being active again" when I was sitting at my desk scratching my head.
21.01.2025 17:54 — 👍 5 🔁 0 💬 1 📌 0
I feel your pain. I had to scavenge some emergency J48s a couple of weeks ago.
16.01.2025 04:35 — 👍 5 🔁 0 💬 0 📌 0
This is beautiful!!! Do you happen to have some scripts for using Microscopy Nodes w/ bpy in python?
15.01.2025 14:15 — 👍 2 🔁 0 💬 1 📌 0
I am a professor in the computer sciences at UW-Madison. My technical interests in trustworthy ML, formal methods, and security.
My other interests are Indian classical music, mindfulness, tennis, and pickleball.
Evangelist for the study of evolution in action by everyone.
Pitt Prof | EvolvingSTEM | biofilms | EvMed | genomics entrepreneur (@SeqCoast.bsky.social, @midauthorbio.bsky.social) | ASM volunteer | lifelong exercise addict ~ swim bike run
Flatiron Research Fellow @flatironinst. Studying temperature sensing ion channels with molecular dynamics and cryoEM❄️🔬.
@sydney_physics allum. he/him
Reproducible bugs are candies 🍭🍬
Machine Learning in Structural Biology workshop co-located with NeurIPS 2025. Dec 6 or 7. San Diego, CA
http://mlsb.io
Still straining against the 2nd law of thermodynamics
*Views are my own
📖Editor at BMC Biology @bmc.springernature.com
🦠Part-time community college prof
🧘🏻♀️200 hour RYT
📍 Philadelphia
Past:
🔬Postdoc Thomas Jefferson University
🧬PhD @stonybrooku.bsky.social
🧪BS @mnstatemankato.bsky.social
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
developmental biologist, fruit fly enthusiast | Wellesley College alumna | Leading Edge Fellow | she/her 🏳️🌈| views and opinions = mine
Official account of the Deep-Sea Biology Society | #DSBSoc
A society for student, academic and professional deep-sea biologists. 🌊
🔗 @dsbsoc-arts.bsky.social
🔗 dsbsoc.org
Official account of the Max Planck Society. Devoted to basic #research in #physics #astronomy #chemistry #biology #earthsciences #materialscience #mathematics #socialsciences and the #humanities; Imprint: https://www.mpg.de/imprint
The American Ornithological Society (AOS) is a global network of professionals working together to advance the scientific study and conservation of birds.
The official account of the American Arachnological Society (AAS). AAS furthers the study and appreciation of arachnids, and publishes the Journal of Arachnology. 🕸️🕷️🦂 & other wonderful #arachnids!
https://linktr.ee/american_arachnology
Die DGN ist die Stimme der Neurologie. Wir fördern Wissenschaft, Forschung, Lehre sowie Fort- und Weiterbildung in der Neurologie. Wir prüfen die wissenschaftliche Debatte und beteiligen uns an der gesundheitspolitischen Diskussion.
Official profile of the European Society for Evolutionary Biology (ESEB) - an international academic society supporting the study of #evolution
https://eseb.org
Publisher of @jevbio.bsky.social
Co-publisher of @evolletters.bsky.social
The SMB is dedicated to the advancement of research at the interface between the mathematical and biological sciences. smb.org
The German Society for Mammalian Biology (Deutsche Gesellschaft für Säugetierkunde, e.V./DGS) promotes research from all fields of mammalogy since 1926.
https://www.mammalian-biology.de
Host of Mammalian Biology https://link.springer.com/journal/42991
Advancing conservation knowledge and practice in Oceania.
https://www.scboceania.org
https://linktr.ee/scboceania
This is the official account of the Society of Island Biology (SIB). For more information go to https://islandbiology.com/.