Super cool work - congratulations @umbislupo.bsky.social & team!
16.01.2026 17:19 — 👍 1 🔁 0 💬 0 📌 0
Congratulations @chaitjo.bsky.social ! 🙌
03.12.2025 19:54 — 👍 1 🔁 0 💬 0 📌 0

RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
15.08.2025 13:26 — 👍 54 🔁 19 💬 0 📌 0
This paper represents a great effort by @roman-bushuiev.bsky.social and his brother @anton-bushuiev.bsky.social. The DreaMS foundation model for mass spectra of small molecules now opens lots of avenues for possible downstream applications. It might be a game changer for computational metabolomics.
24.05.2025 08:21 — 👍 52 🔁 20 💬 0 📌 1
Very nice, thoughtful post - I really enjoyed the read @pascalnotin.bsky.social
10.05.2025 22:16 — 👍 3 🔁 0 💬 0 📌 0
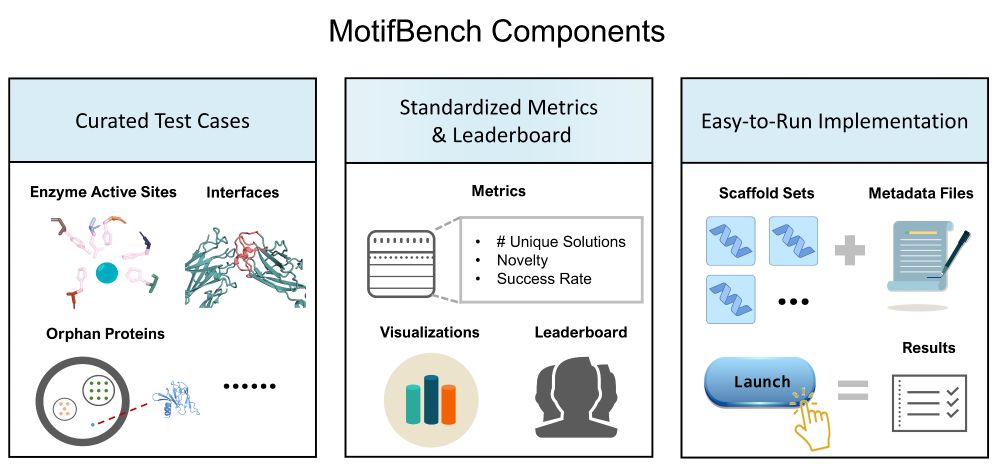
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
19.02.2025 20:49 — 👍 41 🔁 17 💬 1 📌 5
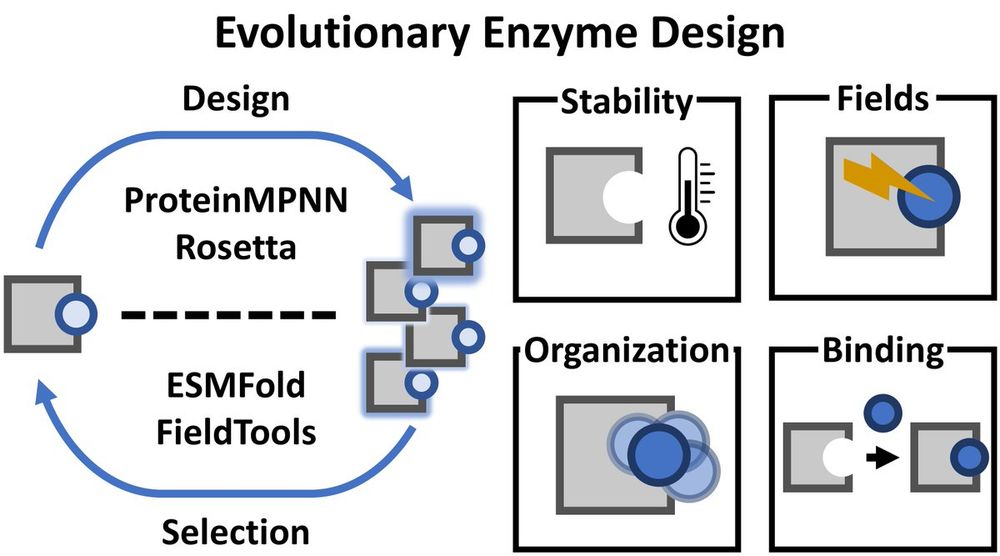
🚨Preprint Alert!🚨
#ProteinDesign is advancing rapidly—wouldn't it be great to seamlessly combine design tools to achieve more than what each can do alone?🤔
Here, we introduce AI.zymes: A modular platform for evolutionary #EnzymeDesign.♻️🖥️
biorxiv.org/content/10.1...
1/🧵
12.02.2025 19:01 — 👍 30 🔁 7 💬 1 📌 1
Amazing, congratulations @machine.learning.bio
23.12.2024 23:02 — 👍 8 🔁 0 💬 0 📌 0
You may have seen a recent pre-print [1] from Jain et al. with strongly worded claims against the experimental results in our DiffDock paper [2]. We initially declined to respond as we saw that this preprint contained falsehoods, misleading comparisons, seemingly deliberate omissions, ...1/n
08.12.2024 21:37 — 👍 30 🔁 11 💬 2 📌 7

Can AI improve the current state of molecular simulation? (Corin & Ari Wagen, Ep #1)
2.1 hours listening time
Can AI improve the current state of molecular simulation?
www.owlposting.com/p/can-ai-imp...
in my first podcast, I spend 2 hours interviewing Corin Wagen and Ari Wagen, two brothers who are building the next generation of molecular simulation for drug discovery and material science
04.12.2024 01:50 — 👍 56 🔁 14 💬 1 📌 3
Thank you for the feedback! That’s great to hear 🙌
04.12.2024 23:44 — 👍 1 🔁 0 💬 0 📌 0
(2/2) ... say a diffusion trajectory then it's doing something you cannot achieve by rsyncing folders (or only *very* cumbersomely). I mostly use it to debug & sanity check my code whilst developing for example
02.12.2024 23:00 — 👍 0 🔁 0 💬 0 📌 0
(1/2) Good question! If you only use it to look at static pymol files that you saved out then yes, it's an alternative to rsyncing your hpc folder. If you use it to for example visualize in-RAM objects during code execution / debugging, or if you use it to manually dock something midway through ...
02.12.2024 22:58 — 👍 0 🔁 0 💬 1 📌 0
Thank you Greg! I actually found out about this functionality by using your fantastic RDKit package and very much based it on your RPC implementation there ( 😉 see the 3. Credits section). I mainly added functionality to send back and forth application states and a wrapper command for ease of use
30.11.2024 08:33 — 👍 0 🔁 0 💬 1 📌 0

Cursor
Built to make you extraordinarily productive, Cursor is the best way to code with AI.
Haha great point ^^ your milage varies but I've found Claude sonnet 3.5 via the cursor.com integration to work reasonably well -- the copilots then pick up patterns if there's already some sensible commands within the context
30.11.2024 08:28 — 👍 1 🔁 0 💬 1 📌 0
For some more guidance on how to use this, Martin Buttenschön wrote a nice blogpost: www.blopig.com/blog/2024/11...
29.11.2024 10:26 — 👍 18 🔁 4 💬 1 📌 1
Check out our MassSpecGym dataset on @polarishq.bsky.social. 🤩
27.11.2024 08:18 — 👍 10 🔁 3 💬 0 📌 0
Conformational dynamics smoothen the fitness landscapes of enzymes. Who knew? 🧪🧶
27.11.2024 04:17 — 👍 31 🔁 6 💬 2 📌 1
Congratulations! This is very exciting news
26.11.2024 22:20 — 👍 1 🔁 0 💬 0 📌 0

Cradle – Cradle raises $73M Series B to Put AI-Powered Protein Engineering in Every Lab
News, updates, tutorials, and more from the makers of Cradle
✨ wheeeeee we raised a series B
AI-powered automated protein optimization: matches or exceeds human performance, works without human intervention. It's pretty cool :D
(PS if you're seriously good at ML eng then we're hiring)
www.cradle.bio/blog/series-b
26.11.2024 08:50 — 👍 81 🔁 3 💬 2 📌 0
Amazing! Congratulations to you and the cradle team 🙌
26.11.2024 22:10 — 👍 1 🔁 0 💬 0 📌 0
Haha where can I order an edition of this? :D
26.11.2024 22:06 — 👍 1 🔁 0 💬 0 📌 0

As added bonus this allows you to use github copilot or cursor's copilot directly in pymol
25.11.2024 15:12 — 👍 0 🔁 0 💬 1 📌 0
My workflow:
I use GPUs on my university's cluster to run models, etc.
When I want to look at my designs, I open a pymol session on my laptop, log into the uni vpn and send the structures to my local remote from an interactive session on the cluster.
25.11.2024 14:50 — 👍 1 🔁 0 💬 1 📌 0
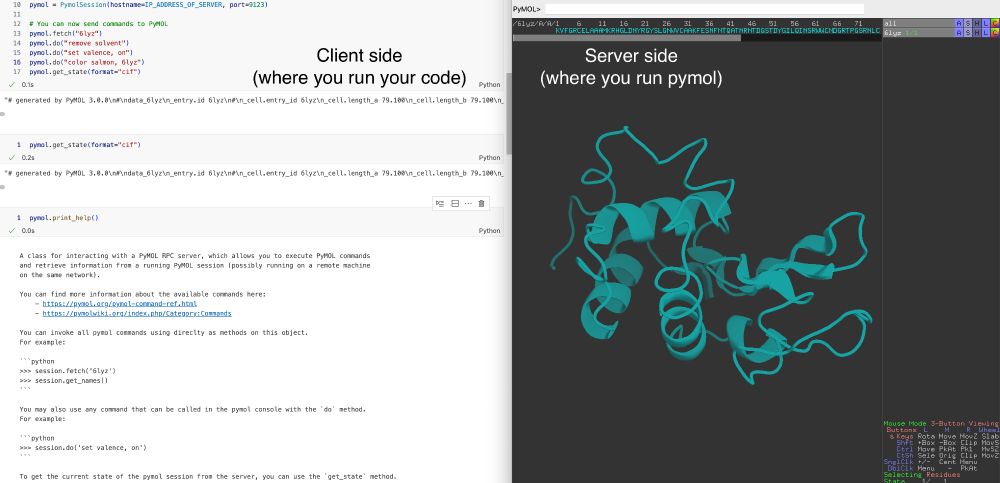
A weekend project from a while back -- this little package (with no dependencies) allows you to interact with pymol remotely.
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
25.11.2024 14:50 — 👍 47 🔁 13 💬 4 📌 3
Done- and hey Linna ((:
21.11.2024 22:28 — 👍 1 🔁 0 💬 0 📌 0
Done ✅ 🙌
21.11.2024 16:47 — 👍 0 🔁 0 💬 0 📌 0
MIT biology lab using computational and experimental methods to study protein structure, function, and interactions
Bear blog of Andrej Karpathy, AI researcher. Started March 2025. ...
🌉 bridged from 🌐 https://karpathy.bearblog.dev/: https://fed.brid.gy/web/karpathy.bearblog.dev
Automated discovery of AI x Bio preprint papers.
Building personalized Bluesky feeds for academics! Pin Paper Skygest, which serves posts about papers from accounts you're following: https://bsky.app/profile/paper-feed.bsky.social/feed/preprintdigest. By @sjgreenwood.bsky.social and @nkgarg.bsky.social
EurIPS is a community-organized, NeurIPS-endorsed conference in Copenhagen where you can present papers accepted at @neuripsconf.bsky.social
eurips.cc
Frontier models for all molecules of life.
Protein biophysicist working on intrinsically disordered proteins in neuroscience and biotech. Assoc. prof at @MolBiolAU and @PromemoAU. (He/Him)
Professor of Theoretical Chemistry @sorbonne-universite.fr & Director @lct-umr7616.bsky.social| Co-Founder & CSO @qubit-pharma.bsky.social | (My Views)
https://piquemalresearch.com | https://tinker-hp.org
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
Achira is building atomistic foundation simulation models to power the future of drug discovery.
http://achira.ai
Scientist and Group Leader of the Simons Machine Learning Center
@SEMC_NYSBC. Co-founder and CEO of http://OpenProtein.AI. Opinions are my own.
Assistant Professor at Dartmouth College
Computational Biophysics / Disordered Proteins / Molecular Recognition
https://www.cameo3d.org/
CAMEO (Continuous Automated Model EvalutatiOn) is a community project to continuously evaluate the accuracy and reliability of 3D complex structure predictions.
Developed by @sib.swiss and @biozentrum.unibas.ch.
Stanford CS PhD student working on ML/AI for genomics with @anshulkundaje.bsky.social
austintwang.com
PhD candidate Hubrecht Institute | Using de-novo protein design to study epigenetics in the Sahtoe lab
ML Scientist @ Prescient Design (Genentech), AI for Drug Discovery
Advancing the frontiers of basic science through grantmaking, research and public engagement. Sign up for our newsletter: simonsfoundation.org/newsletter
Assistant Professor @ École Polytechnique, IP Paris |
Interested in GNNs and Graph Representation Learning |
Website: johanneslutzeyer.com
Working on evaluation of AI models (via human and AI feedback) | PhD candidate @cst.cam.ac.uk
Web: https://arduin.io
Github: https://github.com/rdnfn
Latest project: https://app.feedbackforensics.com











