
New job openings @openprotein.bsky.social across protein foundation model research, computational protein design, and cloud platform engineering
www.openprotein.ai/careers
@tbepler.bsky.social
Scientist and Group Leader of the Simons Machine Learning Center @SEMC_NYSBC. Co-founder and CEO of http://OpenProtein.AI. Opinions are my own.

New job openings @openprotein.bsky.social across protein foundation model research, computational protein design, and cloud platform engineering
www.openprotein.ai/careers

Our preprint on sequence-to-property learning and zero-shot fitness prediction with PoET-2 is live: arxiv.org/abs/2508.04724
PoET-2 is also open sourced on github: github.com/OpenProteinA...
Thanks to the @openprotein.bsky.social team!
Boltz-1 & Boltz-2 now live via GUI & APIs! Predict protein, protein–RNA/DNA/ligand structures with confidence scores & binding affinity metrics for virtual screening. Compare finetuned models in the new overview page to find your best performer fast.
www.openprotein.ai/early-access...
Product update: Indel Analysis lets you score insertions/deletions across your sequence using PoET-2. You can now also compare multiple 3D structures in Mol* to evaluate design alternatives.
Sign up now: www.openprotein.ai/early-access...
How would you use a tool like this? Do you design or screen indels in your work? 4/4
20.06.2025 04:50 — 👍 0 🔁 0 💬 0 📌 0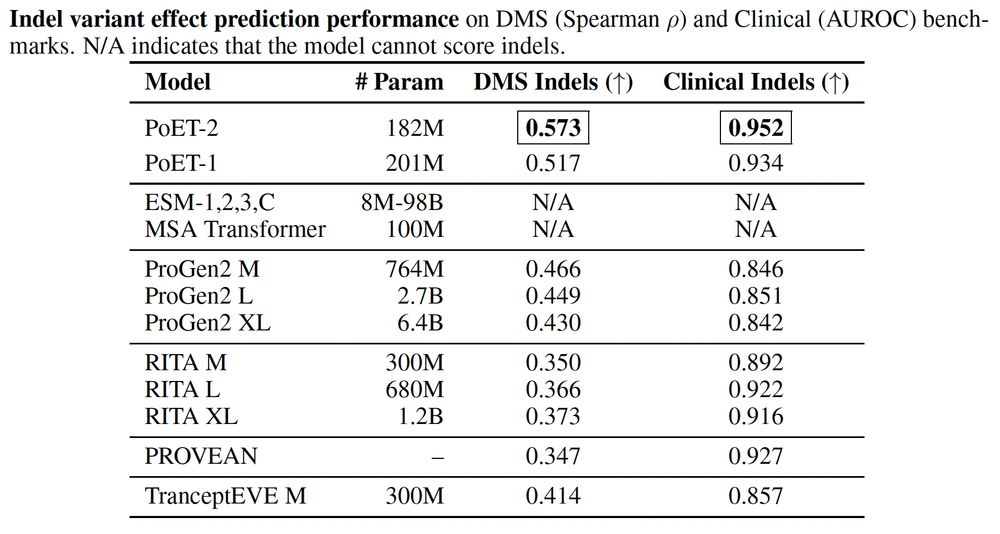
PoET-2 is state-of-the-art for zero-shot variant effect prediction of both DMS indels and clinical indels in ProteinGym.
Indels are still a major challenge for variant effect prediction and protein design. PoET-2 has significantly improved the state-of-the-art for functional and clinical indel variant effect prediction. 3/4
20.06.2025 04:50 — 👍 0 🔁 0 💬 1 📌 0It supports screening deletions, insertion sites, and replacement sites. Explore viable shortened proteins, or insert new structural or functional sequences like localization signals or structural tags. 2/4
20.06.2025 04:50 — 👍 0 🔁 0 💬 1 📌 0Why does no one in AI protein engineering work on indels?
We’re solving this at OpenProtein.AI. Check out our upcoming indel design tool! 🤩 1/4
@openprotein.bsky.social
Have we hit a "scaling wall" for protein language models? 🤔 Our latest ProteinGym v1.3 release suggests that for zero-shot fitness prediction, simply making pLMs bigger isn't better beyond 1-4B parameters. The winning strategy? Combining MSAs & structure in multimodal models!
08.05.2025 00:29 — 👍 25 🔁 7 💬 1 📌 2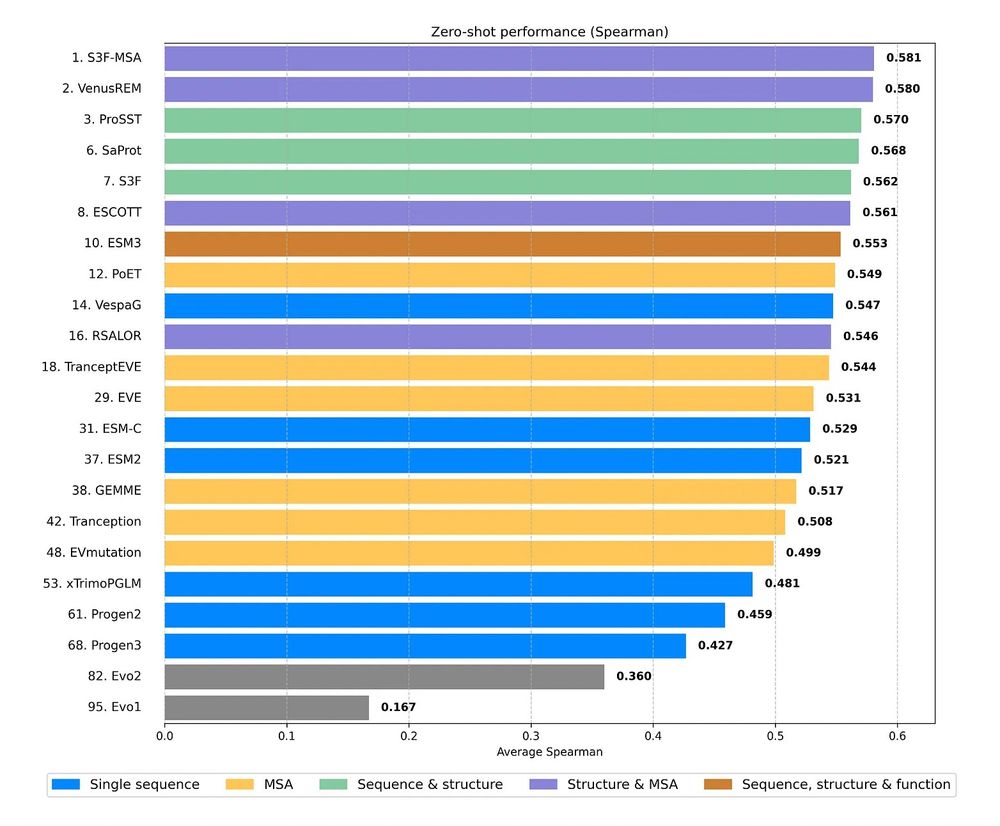
Great to see this comparison with genome language models. The hype around these models seems to have strongly outstripped where they actually are in comparison with protein models.
14.05.2025 03:22 — 👍 1 🔁 0 💬 0 📌 0Product update: PoET-2 now supports structure inputs for enhanced prediction and design via Python APIs. Check out our new inverse folding tutorial to see it in action.
🔗 docs.openprotein.ai/walkthroughs...
Sign up for OpenProtein.AI: www.openprotein.ai/early-access...
Huge thanks to the @openprotein.bsky.social team! We've got more exciting PoET-2 updates to come 🚀
12.05.2025 16:17 — 👍 0 🔁 0 💬 0 📌 0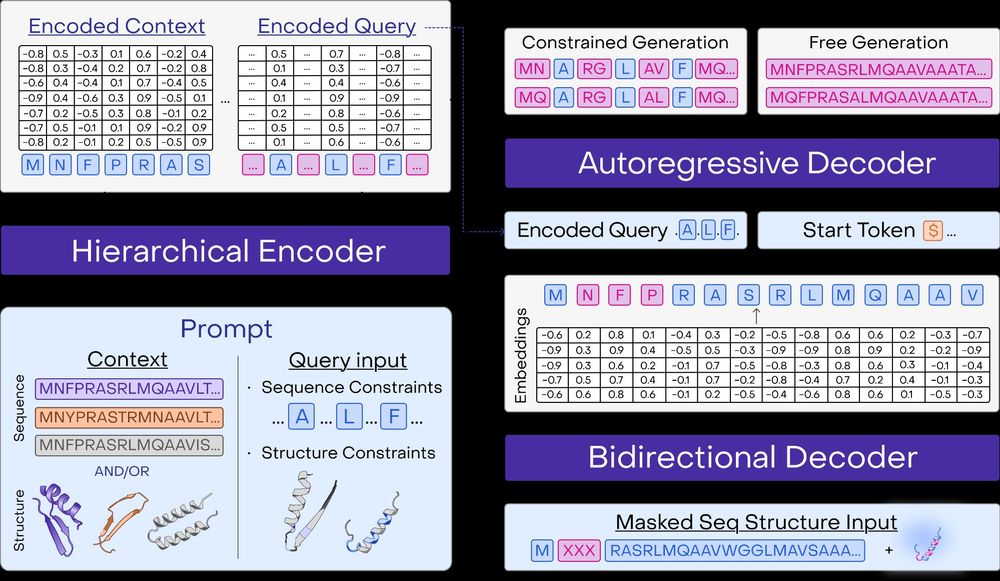
Learn more about PoET-2 in our whitepaper: www.openprotein.ai/a-multimodal...
12.05.2025 16:17 — 👍 1 🔁 0 💬 1 📌 0
Sign up for OpenProtein.AI (free for academic use): www.openprotein.ai/early-access...
and install the python client to get started: github.com/OpenProteinA...
Generative protein sequence design, variant effect prediction, and fine-tuning are now fully supported for PoET-2 with structure and sequence prompts in the @openprotein.bsky.social python client and APIs!
Check out our new walkthrough on inverse folding: docs.openprotein.ai/walkthroughs...

🧬 Protein Revolution: The Tiny Model Making a Massive Impact!
PoET-2 is changing the game in computational protein design, slashing experimental data needs by 30x! 🚀
learn more: www.synbiobeta.com/read/protein...
#ProteinDesign #BiotechInnovation #AIRevolution
Huge thanks to our incredible team @openprotein.bsky.social, especially Tim Truong. This is just the beginning of AI systems that truly understand protein biology.
I can’t wait to see what the community can do with these models! 13/13
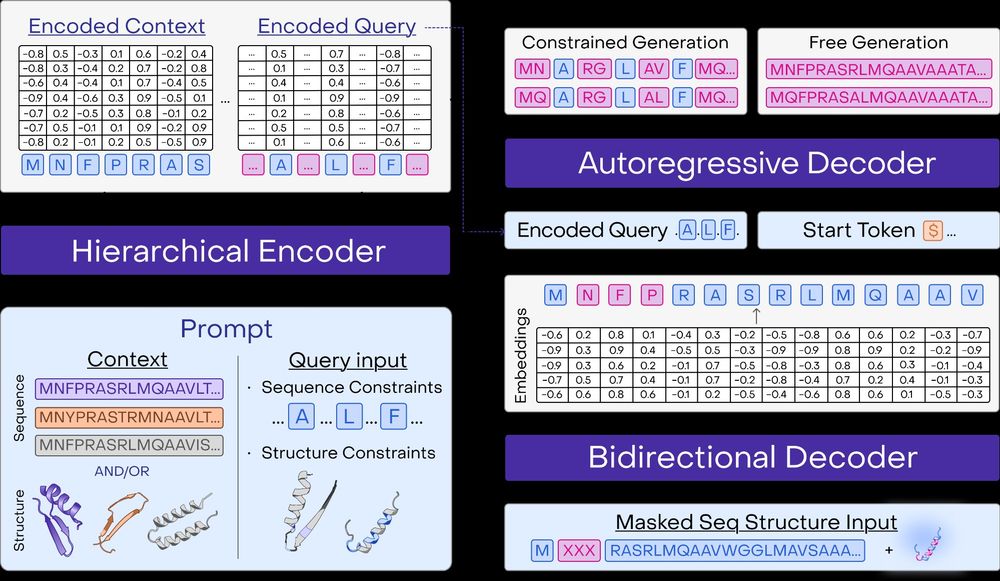
Read more in our white paper: www.openprotein.ai/a-multimodal...
12/13
This has huge implications for protein engineering - from more efficient directed evolution and multiproperty optimization to de novo protein design. 11/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0Most importantly, PoET-2 gets us closer to understanding the sequence-structure-function relationship - learning from just a handful of examples to predict properties and design new sequences. 10/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0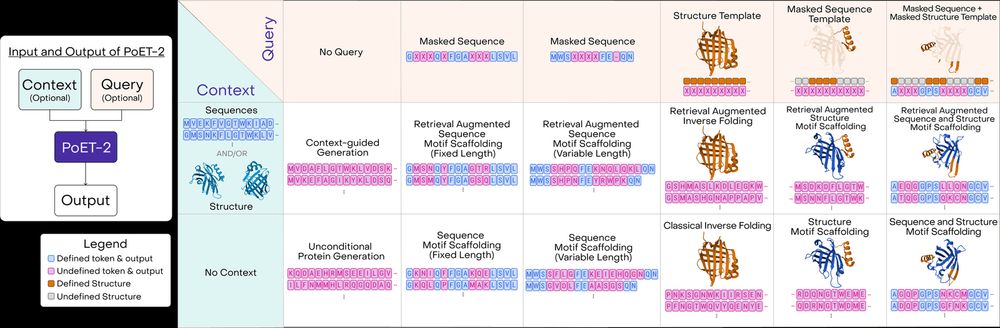
Beyond predictions, PoET-2 introduces a powerful prompt grammar for protein generation. One model for: free sequence generation, inverse folding, motif scaffolding, and more! 9/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0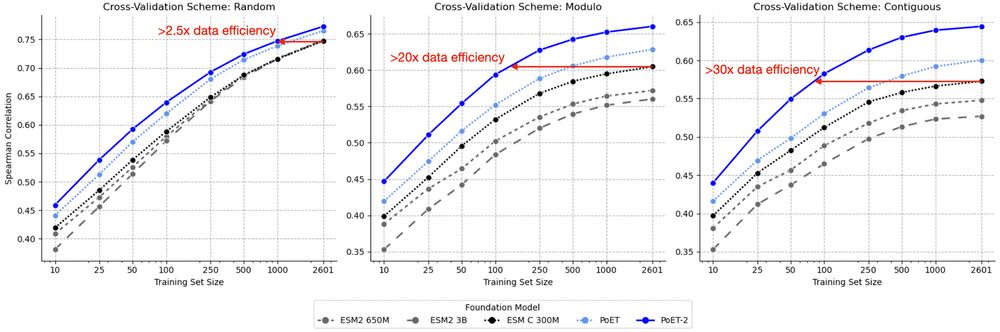
The results show PoET-2 has learned fundamental principles:
* Improves sequence and structure understanding
* Accurate zero-shot function prediction, especially for insertions and deletions
* 30x less data needed for transfer learning
8/13
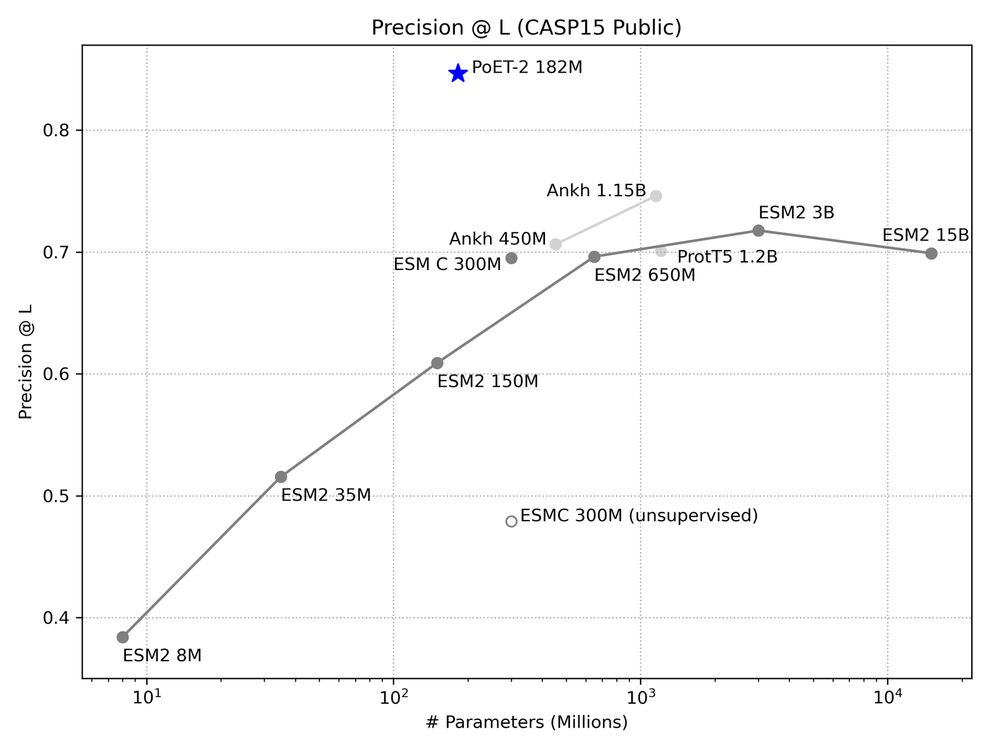
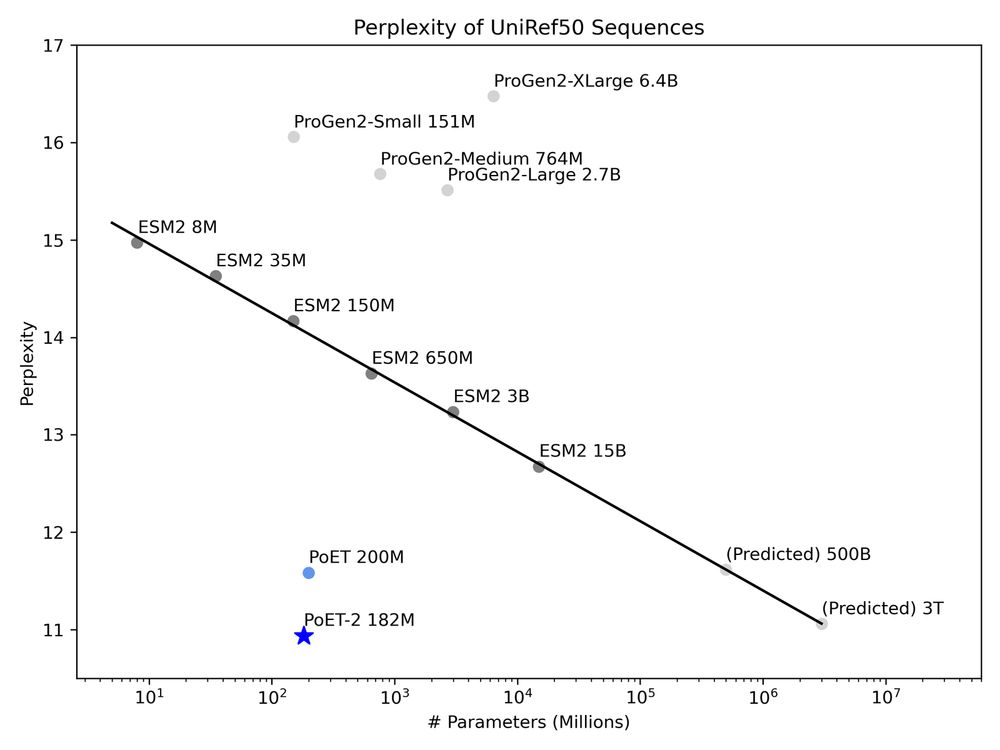
This lets us break conventional scaling laws. PoET-2 achieves with 182M parameters what would require trillion-parameter models using standard architectures. 7/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0Enabled by our tiered attention structure, PoET-2 processes sequence families with order equivariance while preserving long-range dependencies within and between sequences, enabling processing of large sequence families with optional structural data. 6/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0Rather than regurgitating databases, PoET-2 "meta-learns" evolutionary principles through in-context learning - inferring structural and functional constraints at inference time from small numbers of examples. 5/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0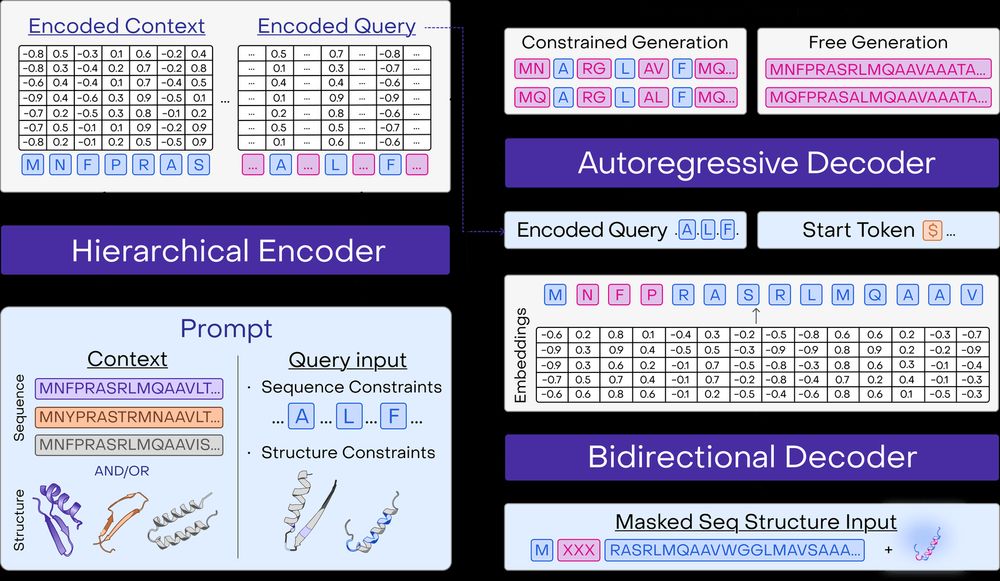
PoET-2 takes a different approach. Instead of massive scale, we developed a multimodal architecture that learns to reason about sequences, structures, and evolutionary relationships simultaneously. 4/13
11.02.2025 14:30 — 👍 3 🔁 1 💬 1 📌 0But memorizing sequences isn't enough. The real challenge: can we build models that learn the fundamental principles that govern how proteins evolve and function? 3/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0Since our first protein language models in 2019 (in Bonnie's lab!), the field has focused on scale - building ever-larger models up to 100B parameters to extract information from natural sequence databases. 2/13
11.02.2025 14:30 — 👍 1 🔁 0 💬 1 📌 0Excited to share PoET-2, our next breakthrough in protein language modeling. It represents a fundamental shift in how AI learns from evolutionary sequences. 🧵 1/13
11.02.2025 14:30 — 👍 31 🔁 15 💬 1 📌 0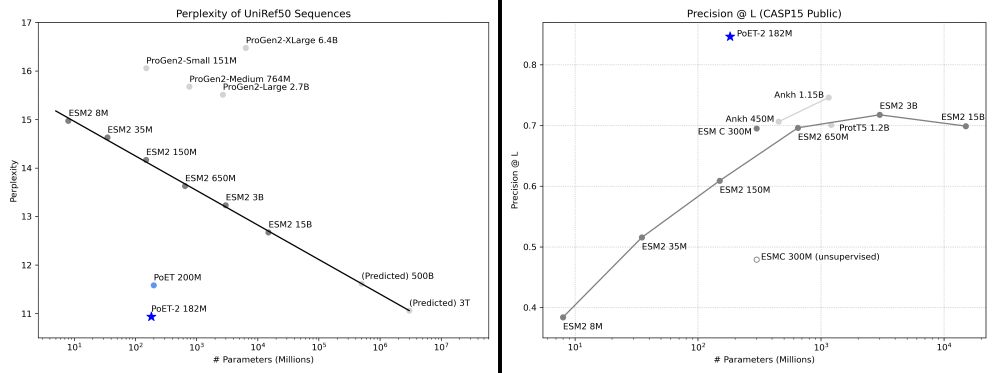
🧬 Announcing PoET-2: A breakthrough protein language model that achieves trillion-parameter performance with just 182M parameters, transforming our ability to understand proteins.
11.02.2025 13:38 — 👍 4 🔁 2 💬 1 📌 0