Special thanks to @maxraas.bsky.social @mfseidl.bsky.social Peter Verrijzer & Berend Snel for all the help and collaboration on this project and to the reviewers and editors for all their helpfull suggestions to improve the paper!
10.11.2023 13:53 — 👍 1 🔁 1 💬 0 📌 0
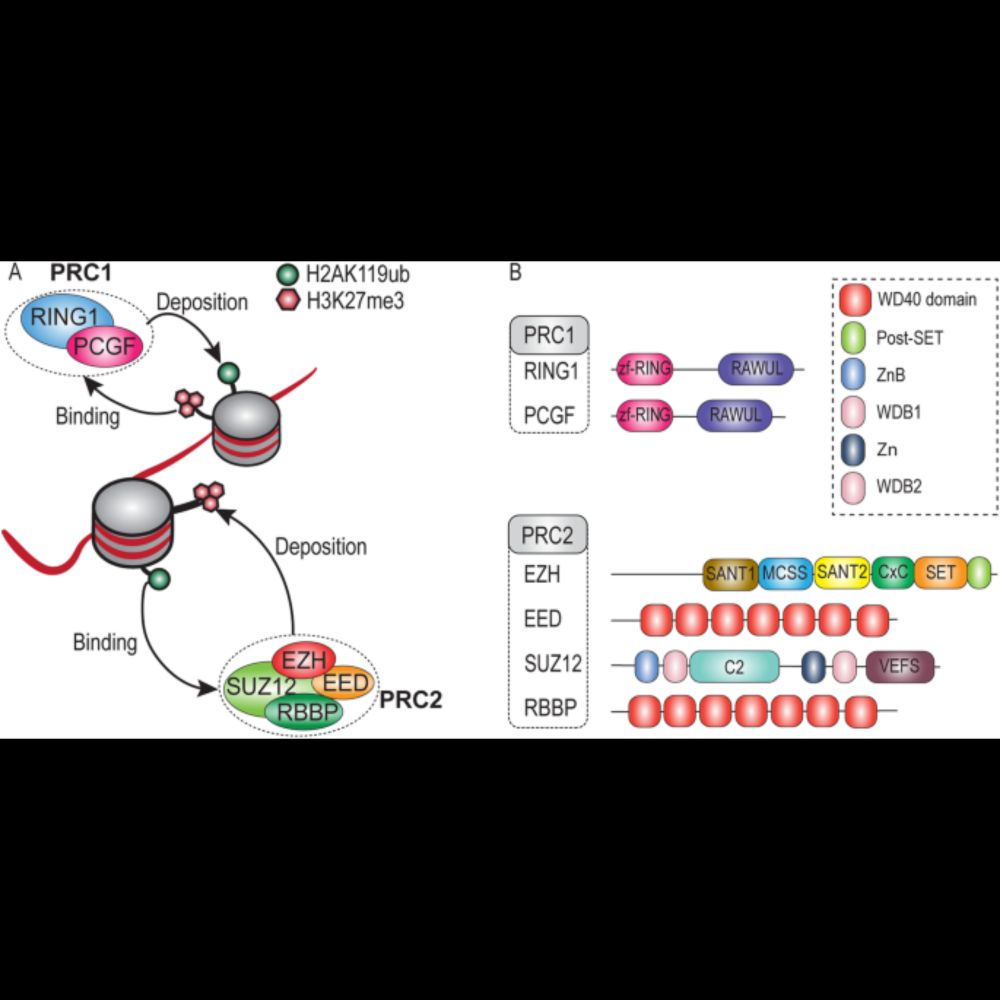
In short, our results demonstrate the independent evolution and function of PRC1 and PRC2, and show that crosstalk between these complexes is a secondary development in evolution.
10.11.2023 13:52 — 👍 0 🔁 0 💬 1 📌 0
Furthermore,the identification of orthologs for ncPRC1-defining subunits in unicellular relatives of animals and of fungi suggests that the origin of ncPRC1 predates that of cPRC1, and we develop a scenario for the evolution of cPRC1 from ncPRC1.
10.11.2023 13:52 — 👍 0 🔁 0 💬 1 📌 0
Strikingly, a substantial portion (42%) of the organisms that contain Polycomb orthologs encode for only PRC1 or only PRC2, showing that their evolution since LECA is largely uncoupled.
10.11.2023 13:52 — 👍 0 🔁 0 💬 1 📌 0
Combining phylogenetics, structural analyses and comparative genomics we determined the deep origin and evolution of the core Polycomb proteins. We found that both PRC1 and PRC2 were present in the Last Eukaryotic Common Ancestor (LECA).
10.11.2023 13:52 — 👍 1 🔁 0 💬 1 📌 0
Protein and coffee lover, father of two, professor of biophysics and sudo scientist at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen 🇩🇰
Engineer/scientist, Nobel Prize in Chemistry 2018
I love evolution, enzymes, protein engineering, AI
Linus Pauling Professor at Caltech
ML for proteins
PhD student at Caltech with Frances Arnold and Yisong Yue
Prev: MSR intern, Berkeley alum
https://www.francescazfl.com/
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
🧪 Research Scientist @nvidia and PhD student @Oxford staring at proteins all-day
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
I do SciML + open source!
🧪 ML+proteins @ http://Cradle.bio
📚 Neural ODEs: http://arxiv.org/abs/2202.02435
🤖 JAX ecosystem: http://github.com/patrick-kidger
🧑💻 Prev. Google, Oxford
📍 Zürich, Switzerland
Generative models for protein design.
Group leader at CRG
Protein Design / ML @ Profluent Bio | Molecular Biophysics PhD @ Johns Hopkins
🇪🇪 De Novo enzyme design • HHMI Postdoc at the Institute for Protein Design at University of Washington
Designed the first apps for Uber.com, Booking.com, Catawiki.com and many others.
Co-founder http://operator.exchange and http://cradle.bio
BRENDA is the comprehensive electronic information resource that comprises molecular and biochemical information on enzymes.
Visit:
https://www.brenda-enzymes.org/
https://hub.dsmz.de/#/
Developing ML methods for Drug Discovery @ Novartis 🧬💻 | prev: PhD student @ University of Edinburgh with Toni Mey
Group Leader at the Institute of Organic Chemistry and Biochemistry (IOCB Prague @iocbprague.bsky.social). Plant biochemistry, metabolomics, genomics, bioML, etc. https://www.pluskal-lab.org
Postdoc at Stanford. Previously PhD student at Berkeley AI research. Trying to understand proteins with microfluidics and machine learning.
www.karlk.net
Computational biochemist (he/him), PI of the http://vanderkampgroup.com. Likes #enzymes & #compchem, #diversity #inclusion
PhD student Uni of Cambridge | Enzyme engineering and design for sustainable biocat | Droplet microfluidics as a tool for mega-scale data collection
PhD @ Cambridge in AI for Bio | Interested in generative modelling for drug discovery and science policy 🇬🇧
Website: cch1999.github.io
Blog: harrisbio.substack.com
Database: harrisbio.notion.site
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique
Researcher at Google DeepMind. Previously Arnold group at Caltech. Making useful molecules!