
Mathematisch-Naturwissenschaftliche Fakultät
Heute ernannte Rektorin Prof. Dr. Anja Steinbeck den Biotechnologen Dr. Johannes Rebelein zum W3-Professor für Nachhaltige Biotechnologie. Rebelein wird damit auch Direktor des Instituts für Molekular...
A childhood dream has come true – I was appointed Full Professor for Sustainable Biotechnology at the University of Düsseldorf @hhu.de : tinyurl.com/f43s74hp and as Director of the Institute for Molecular Enzyme Technology (IMET, www.iet.uni-duesseldorf.de/en/) at FZ Jülich @fz-juelich.de .
06.03.2026 07:23 —
👍 37
🔁 5
💬 1
📌 2

Happy to share our new paper on proton transfer in metalloenzymes, out now in @pccp.rsc.org!
Exploiting crystallography, electrochemistry, FTIR spectroscopy, and QM calculations we identified a Zundel ion in the catalytic proton tranfer pathway of [FeFe]-hydrogenase.
doi.org/10.1039/D5CP...
04.03.2026 12:49 —
👍 12
🔁 5
💬 1
📌 0
New preprint by @alok-bharadwaj.bsky.social explaining the workings of SURFER, a simple tool to segment membrane, micelle or nanodisc densities in cryoEM maps.
Try it via the @chimerax.ucsf.edu ToolShed:
cxtoolshed.rbvi.ucsf.edu/apps/chimera...
Despite LocScale in the name, it works on any map.
27.02.2026 12:17 —
👍 22
🔁 8
💬 0
📌 0
Shades of pink we picked for HDCR core 😁 was beautiful!
20.02.2026 12:11 —
👍 2
🔁 0
💬 0
📌 0
At long last, the Chlamy chloroplast ribosome hat 🎩 (now renamed the arm 💪) is here to say hello👋! We first spotted this extended structure in green algae a decade ago, and @florentwaltz.bsky.social did beautiful work bringing it into focus! #CryoEM ❄️ #TeamTomo 🔬 #PlantScience 🌾 #ProtistsOnSky 🧪
10.02.2026 13:53 —
👍 50
🔁 11
💬 0
📌 0

This is what I call a booster 🚀: Milton and colleagues use a novel, bipyridium-type redox mediator to increase the electrochemical H2 evolution activity of [FeFe]-hydrogenase by a factor of 10 compared to previous protocols.
pubs.acs.org/doi/full/10....
25.01.2026 20:43 —
👍 5
🔁 1
💬 0
📌 1

An incredibly challenging single particle cryo-EM structure. Capturing both the N and C termini was difficult enough, but the project required a major effort to resolve the full length protein structure to capture the overall architecture of the KICSTOR-GATOR1 complex.
Well worth it in the end!
09.01.2026 07:11 —
👍 10
🔁 2
💬 1
📌 3

I am very excited to share our new preprint - "A Zundel Ion in the Catalytic Proton Transfer Pathway of [FeFe]-Hydrogenase" - for which we revisited an older theory of ours, and came to surprising conclusions... 🧵
doi.org/10.26434/che...
06.11.2025 12:40 —
👍 11
🔁 4
💬 1
📌 1
Excited to share our latest @nature.com: How does naloxone (Narcan) stop an opioid overdose? We determined the first GDP-bound μ-opioid receptor–G protein structures and found naloxone traps a novel "latent” state, preventing GDP release and G protein activation.💊🧪 🧵👇 www.nature.com/articles/s41...
05.11.2025 16:22 —
👍 123
🔁 35
💬 6
📌 2
Our latest work on the nitrogenase-like methylthio-alkane reductase, which specifically reduces reduces carbon-sulfide bonds is now out @natcatal.nature.com: doi.org/10.1038/s419.... We find for the first time large #nitrogenase metalloclusters (P- and L-cluster) outside nitrogenases.
23.10.2025 10:09 —
👍 49
🔁 20
💬 4
📌 1
Great work from the lab of @rebeleinlab.bsky.social ! Now out in @natcatal.nature.com
23.10.2025 09:52 —
👍 1
🔁 0
💬 0
📌 0

A diagram showing the environmental distribution of methyl coenzyme M reductase encoding Thermoproteota.
New preprint: Methyl co-enzme M reductase encoding (potentially methanogenic) Thermoproteota are widespread and transcriptionally active in diverse anoxic ecosystems! @dr-zj.bsky.social, Matthew Kollom & @emileyeloe-fadrosh.bsky.social www.biorxiv.org/cgi/content/... Funded by the DOE BER program.
16.10.2025 21:10 —
👍 19
🔁 10
💬 1
📌 0

Locating the missing chlorophylls f in far-red photosystem I
The discovery of chlorophyll f-containing photosystems, with their long-wavelength photochemistry, represented a distinct, low-energy paradigm for oxygenic photosynthesis. Structural studies on chloro...
Have you ever looked at the night sky and wondered if there’s a chlorophyll f in the reaction centre of Photosystem I from far-red light adapted cyanobacteria? 🦠
Well, we did (don’t judge) and the work that followed is now out as a First Release in @science.org
www.science.org/doi/10.1126/...
11.10.2025 10:37 —
👍 64
🔁 19
💬 2
📌 2
Cryo-EM structure of Chlamydomonas Photosystem I complexedwith the alternative electron donor cytochrome c6 www.biorxiv.org/content/10.1101/2025.09.08.674899v1 #cryoEM
09.09.2025 07:10 —
👍 6
🔁 2
💬 1
📌 0
Initial attempt at replicating in relion (parameters in next post). This is for Aca2-RNA, using a 100k subset of the 2D-classified particles (no prior 3D cleanup).
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
26.09.2025 02:28 —
👍 57
🔁 24
💬 1
📌 2
Happy to share that our story on the bacterial archaellum was published today in @natmicrobiol.nature.com
www.nature.com/articles/s41...
Congrats to the authors: @sshamphavi.bsky.social @loumollat.bsky.social @mariejoest.bsky.social Najwa Taib and @sgribaldo.bsky.social
17.09.2025 17:41 —
👍 122
🔁 58
💬 7
📌 8
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
13.09.2025 00:04 —
👍 266
🔁 82
💬 20
📌 14

Sub-cellular chemical mapping in bacteria using correlated cryogenic electron and mass spectrometry imaging
Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social
31.08.2025 18:44 —
👍 142
🔁 62
💬 3
📌 4
PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
It is out now 🥳🥂
I am very happy to be part of this nice collaboration in which we were able to characterize different bacterial chemoreceptors and their ligand specificities. Amazing teamwork with Wenhao Xu, the Sourjik lab, @bangebalcony.bsky.social, et. al 🤩
👇👇👇
www.pnas.org/doi/10.1073/...
26.08.2025 21:06 —
👍 3
🔁 2
💬 0
📌 0
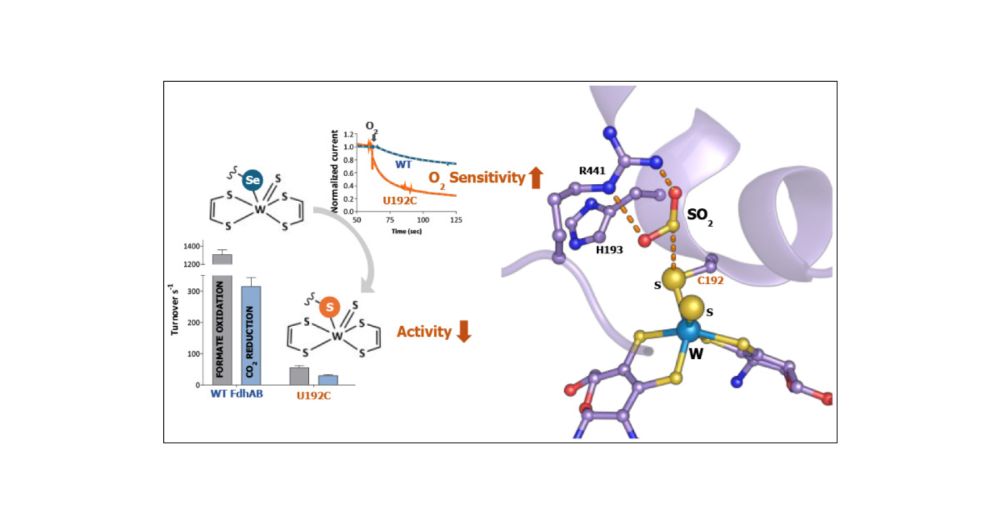
The Role of Selenocysteine in Catalysis and Oxygen Tolerance of a W-Dependent Formate Dehydrogenase
Metal-dependent formate dehydrogenases (FDHs) catalyze, under mild conditions, the reversible reduction of CO2 to formate, a versatile C1 feedstock that can contribute to a carbon-neutral economy. Metal-dependent FDHs are the most widespread selenoproteins found in bacteria, and around 44% of them include selenocysteine (Sec) as a ligand to the Mo/W active site. In the sulfate-reducer Nitratidesulfovibrio vulgaris Hildenborough, the main FDH responsible for CO2 reduction is the W/Sec-dependent FdhAB, which is among the most active CO2 reductases reported so far. In contrast to most metal-dependent FDHs, this enzyme is relatively O2-tolerant and can be purified aerobically. In this work, we evaluated the role of Sec in the catalytic and stability properties of the W/Sec-FdhAB. For that, a Sec-to-Cys variant (U192C) was created, its catalytic and spectroscopic properties were characterized, and its crystal structure was determined. Sec substitution by Cys strongly affects activity, decreases the KM for formate, and increases susceptibility to O2. While Sec-to-Cys replacement induces only weak changes of the WV EPR signals, using 77Se-labeled enzyme, we could show that Sec undoubtedly coordinates the W metal in the WV redox state. The crystal structure of U192C confirmed previous findings on the redox switch mechanism of activation and protection of FdhAB, while revealing a putative catalytic intermediate of FdhAB with Arg441 orienting a CO2 substrate analog (probably SO2) in the active site. Overall, the results indicate that Sec plays a critical role in the high activity displayed by W/Sec-FdhAB, and that it may also be involved in or modulate the proton transfer to and from the active site.
Very excited about this story: Inês Pereira et al. reveal the role of selenocysteine in catalysis and O2-tolerance of a tungsten-dependent formate dehydrogenase (via ACS Catalysis)
doi.org/10.1021/acsc...
19.07.2025 10:46 —
👍 5
🔁 2
💬 0
📌 0

Proud to see our work on metalloenzymes - and methyl coenzyme M reductase (MCR) in particular - featured in the print version of Berlin's @tagesspiegel.de 🤩
⛓️ www.uni-potsdam.de/fileadmin/pr...
17.07.2025 09:09 —
👍 16
🔁 6
💬 0
📌 0

Atomic-resolution structure of a chimeric Powassan tick-borne flavivirus
The 2.8-Å resolution structure is presented for a chimeric construct of an emerging tick-borne flavivirus, Powassan virus.
New in @science.org
We built a safe, atomic-resolution version of the Powassan virus—a deadly tick-borne #flavivirus with no vaccine.
✅ First structure ever
✅ Safe to handle (BSL-2)
✅ Platform for vaccines & antivirals
Let me show you 🧵 (1/5)
Link: www.science.org/doi/10.1126/...
09.07.2025 20:11 —
👍 32
🔁 13
💬 1
📌 0
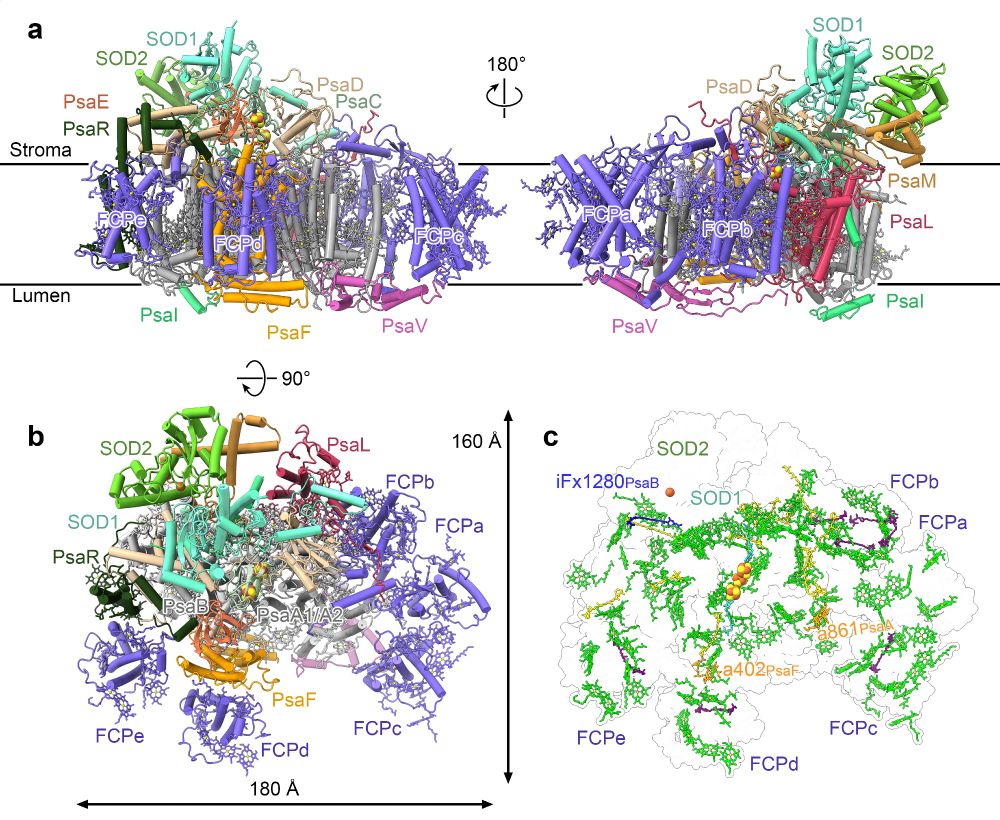
Our new preprint: how photoprotection is spatially coordinated with energy metabolism. Photosystem I is major source of radicals. We report EM structure of PSI in complex with superoxide dismutase SOD: direct link between energy conversion & oxidative stress defence
www.biorxiv.org/content/10.1...
07.07.2025 06:01 —
👍 44
🔁 10
💬 1
📌 1

Our new consensus statement on reducing and reporting contamination in microbiome studies is the cover image of this month's Nature Microbiology: www.nature.com/articles/s41... In the cover photo, you can see our postdoc Sophie Holland in fully PPE sampling the atmosphere of terrestrial Antarctica.
03.07.2025 15:21 —
👍 34
🔁 11
💬 2
📌 0
Happy to share Yiting's new paper on artificial micro hydrogenase aka... Ferredoxin⁉️
Out now in Advanced Science doi.org/10.1002/advs...
19.06.2025 13:38 —
👍 27
🔁 8
💬 0
📌 0

Our ferredoxin story @chemicalscience.rsc.org now in print, just in time for my presentation tomorrow #ISABC17 👏🏻
Sahin et al., Chem. Sci., 2025, 16, 10465-10475
pubs.rsc.org/en/content/a...
15.06.2025 20:56 —
👍 23
🔁 4
💬 1
📌 0
Excellent work from my dear friend Sriram! Many congratulations @georghochberg.bsky.social !
11.06.2025 17:41 —
👍 2
🔁 0
💬 0
📌 0
Excellent work from my colleagues Tristan and Eva on the structure of the ancient sodium pumping Mtr complex from methanogens! Happy to contribute alongside @tomaspascoa.bsky.social @ezimmer.bsky.social @schullerjm.bsky.social
04.06.2025 19:37 —
👍 5
🔁 0
💬 0
📌 0
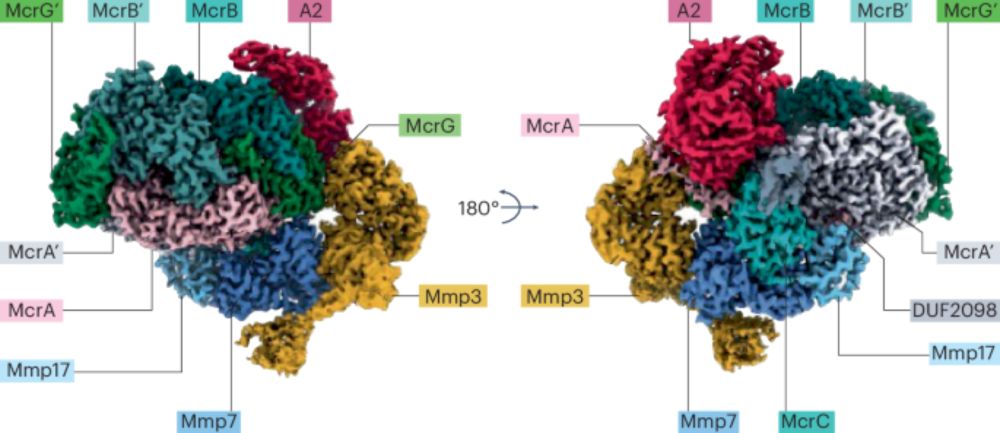
Clustering for methane - Nature Catalysis
Nature Catalysis - Clustering for methane
CLUSTERING FOR METHANE
Nice write-up @natcatal.nature.com of our recent article on MCR activation 🙏🏻
www.nature.com/articles/s41...
04.06.2025 07:15 —
👍 9
🔁 3
💬 0
📌 1

















