What a ride so far… 🎂
08.12.2025 10:28 — 👍 2 🔁 0 💬 0 📌 0David Ochoa
@d0choa.bsky.social
Target discovery | Computational Biology | Human Disease Genetics | ML | Cloud computing - @opentargets.org - @ebi.embl.org
@d0choa.bsky.social
Target discovery | Computational Biology | Human Disease Genetics | ML | Cloud computing - @opentargets.org - @ebi.embl.org
What a ride so far… 🎂
08.12.2025 10:28 — 👍 2 🔁 0 💬 0 📌 0
A project many years in the process, we’re pleased to present our work on multi-ancestry meta-analysis across a boatload of traits in the UK Biobank: www.nature.com/articles/s41...
18.09.2025 17:25 — 👍 63 🔁 25 💬 1 📌 0If you care about regulatory variants don’t miss the E2G extension of the @opentargets.org Platform. There are synergies that only a fully open science allows and this is a good one!
18.09.2025 17:06 — 👍 11 🔁 1 💬 0 📌 0
🚨New preprint just dropped 🚨
medrxiv.org/content/10.1101/2025.06.24.25330216
The main output from my PhD is finally public and we’re SUPER excited about the findings! If you’re interested in what we learnt about IBD with a massive 700+ sample sc-eQTL dataset of the gut, read on!

📅 Join our Q&A drop-in on June 19 at 3pm BST to explore the new features and ask questions:
🔗 www.linkedin.com/events/25-06...
📰 Read more on our blog:
🔗 blog.opentargets.org/open-targets...
Thanks @helenacornu.bsky.social for the infographic!
Every update helps complete the puzzle 🧩 of target discovery.
Huge thanks to our data providers, partners, and the entire Open Targets team for their tireless work 🙏
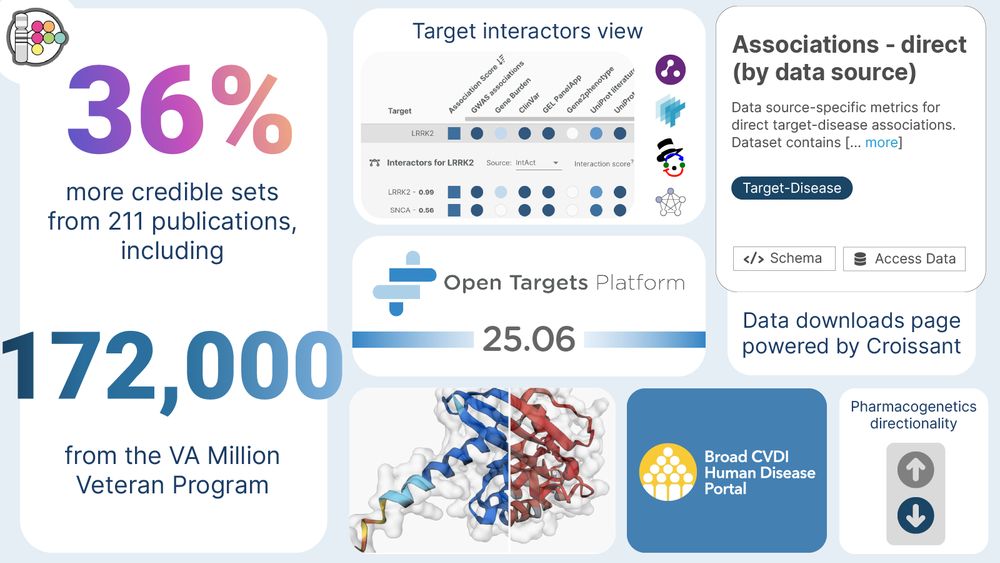
🥐 We’ve also redesigned the data downloads section — now built on the ML Commons Croissant standard.
Better documentation + improved AI-readiness = easier use for researchers and developers alike.
More highlights from the release:
🧬 New gene burden studies
💊 Expanded pharmacogenetics annotations
🧠 Interacting protein evidence from IntAct, Reactome, SIGNOR, STRING
🔬 First look at our molecular structure viewers!
Over the past 3 months, we’ve added GWAS from:
VA Million Veteran Program
210+ publications
Including the Nature meta-analysis of osteoarthritis (April 2024) 🧠
All integrated + contextualised with existing data + functional genomics.
🧬 One of our big promises with bringing Open Targets Genetics to the Platform was keeping post-GWAS analysis fresh and integrated.
This release brings a 36% increase in GWAS credible sets, thanks to collaboration with the @gwascatalog.bsky.social and enhanced pipelines.
🚀 The Summer ☀️ @opentargets.org Platform release is here!
If you thought the last update was big… just wait till you see what’s inside.
A quick thread on the highlights 👇
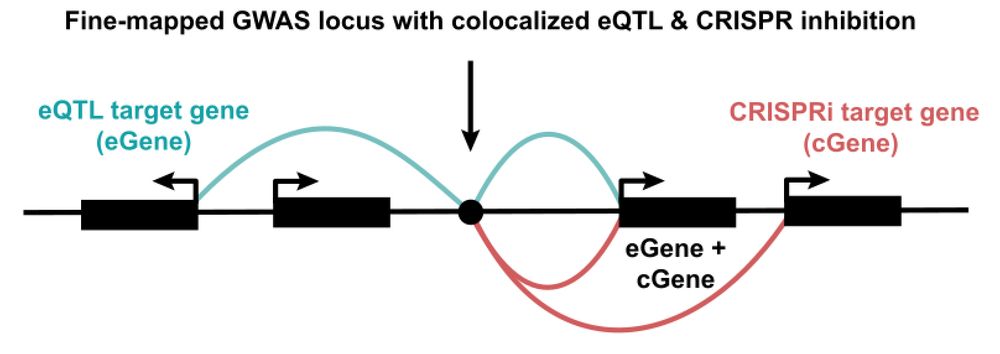
Our new contribution to the quest to find causal GWAS genes! Sam Ghatan from my lab at @nygenome.org led a systematic comparison of eQTLs and CRISPRi+scRNA-seq screens. TL;DR: they provide highly complementary insights, with ortogonal pros and cons. 🧵👇
www.biorxiv.org/content/10.1...

The most famous heuristic in mapping gwas snps to genes is "it's usually the closest gene".
But only slightly less well-known is this: consider the colocalized phenotypes.
That is, a genetic variant seldom disrupts exactly one phenotype.
What else does tugging on that thread do?
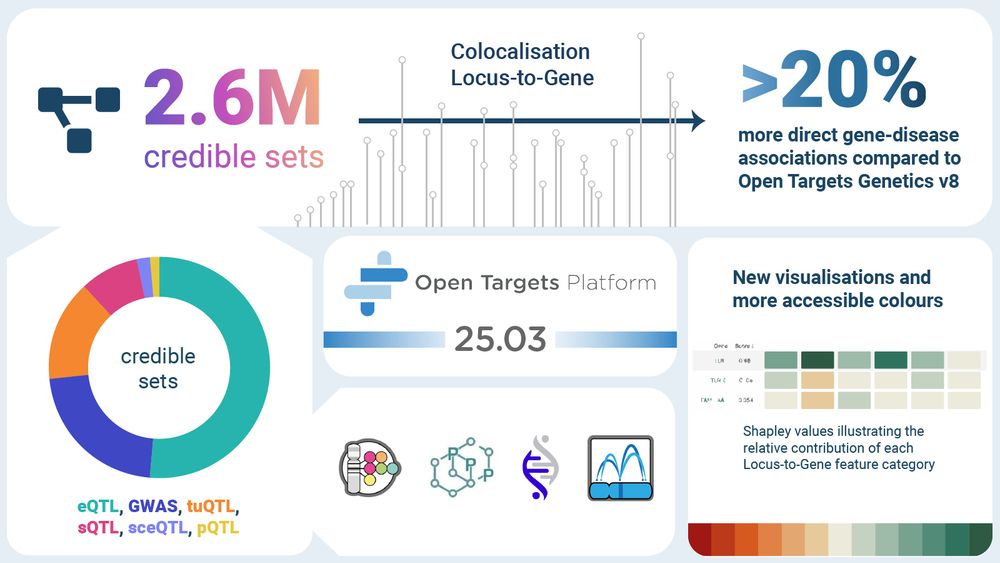
Big thanks to the community for the great feedback we are receiving on the spring @opentargets.org Platform release. There are many positive and new ideas for improving our scientific interpretation and products. Feedback is a critical aspect of an open project's lifecycle. Please keep it coming 🧬🖥️
28.03.2025 11:18 — 👍 9 🔁 4 💬 0 📌 0🚀 Big news! We've just published the official guidelines for submitting affinity proteomics data to PRIDE @pride-ebi.bsky.social (supported technologies Olink & SomaScan)!
Get ahead of the curve—check them out & start your submissions! 👇
🔗 github.com/PRIDE-Archiv...
#Proteomics #Olink #SomaScan
The @opentargets.org Platform Spring 🌼 release brings a step-change in how we address common disease genetics. We included the results of a large-scale analysis on GWAS and functional genomics studies to inform target selection further 🧬 👩💻
blog.opentargets.org/a-step-chang...
We might not get everything right from the first shot. Still, we hope that by providing an open framework and the community's help, we could consolidate our collective understanding of disease-causing genetics. Stay tuned!
17.12.2024 17:06 — 👍 1 🔁 0 💬 0 📌 0The merged product will bring the best of both worlds in a single web application, covering all journeys from identifying the likely causal signals to the prioritising factors relevant to target progression
17.12.2024 17:06 — 👍 1 🔁 0 💬 1 📌 0This update will power our current target identification coverage, as well as the mechanistic interpretation and biological context
17.12.2024 17:06 — 👍 0 🔁 0 💬 1 📌 0The update will include a refreshed post-GWAS analysis covering state-of-the-art data and methodologies, resulting on 2.5M GWAS and molQTL credible sets, colocalisation analysis and L2G predictions
17.12.2024 17:06 — 👍 1 🔁 0 💬 1 📌 0From next spring, the Open Targets Platform will incorporate the best of Open Targets Genetics into an integrated drug discovery platform 🧵
17.12.2024 17:06 — 👍 18 🔁 7 💬 2 📌 0
What do GWAS and rare variant burden tests discover, and why?
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
Specificity, length, and luck: How genes are prioritized by rare and common variant association studies https://www.biorxiv.org/content/10.1101/2024.12.12.628073v1
16.12.2024 10:33 — 👍 46 🔁 24 💬 0 📌 1Out now! A look back at all the changes to the Open Targets Platform in the past two years 🖥️🧬
We've focused on adding data and analyses to help build therapeutic hypotheses, from expanding our associations page...
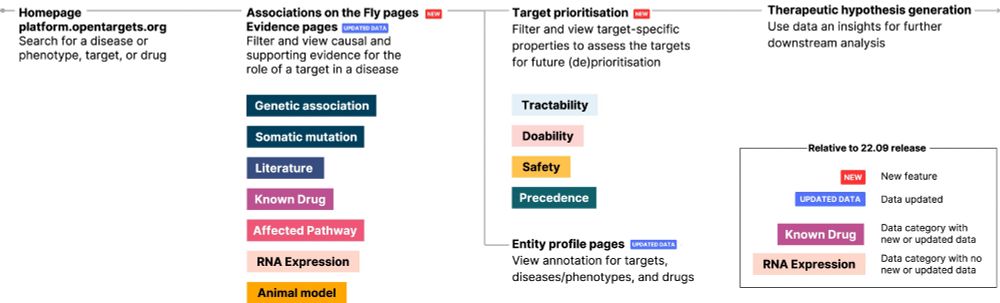
In the upcoming NAR issue, we summarise the last 2 years of updates in the @opentargets.org Platform. One step at a time... academic.oup.com/nar/advance-...
06.12.2024 17:06 — 👍 15 🔁 8 💬 1 📌 0
Instarted making a list the other day, with the help of others adding their own faves: github.com/MichelNivard...
02.12.2024 20:57 — 👍 9 🔁 4 💬 1 📌 1
crispy morning in the genome campus…
28.11.2024 13:27 — 👍 8 🔁 0 💬 0 📌 0plain and simple 👌
22.11.2024 09:34 — 👍 5 🔁 0 💬 0 📌 0



Moving day at @opentargets.org It always amazed me the UK ability to appreciate remarkable scientists. The new @ebi.embl.org building couldn’t be named after a more inspirational figure
21.11.2024 20:01 — 👍 15 🔁 0 💬 1 📌 0My group's work dissecting the contribution of common variants to rare neurodevelopmental conditions is now out at nature.com/articles/s41..., led by co-first authors Qinqin Huang (not yet on blue sky) and @emiliewigdor.bsky.social . See below for Emilie's tweetorial.
20.11.2024 16:15 — 👍 122 🔁 42 💬 7 📌 6