
BioModelAnalyzer Logo
This model can then be further edited and interrogated using the BioModelAnalyzer GUI (biomodelanalyzer.org) and command-line tools (github.com/hallba/BioMo...).
26.05.2025 21:49 — 👍 4 🔁 0 💬 1 📌 0

Network
visualization depicting model fitting and error distribution.
MAGELLAN allows the generation of a computational model from pathway databases, and the fitting of these models to a user-specified specification, resulting in a transparent and interpretable model.
26.05.2025 21:49 — 👍 1 🔁 0 💬 1 📌 0
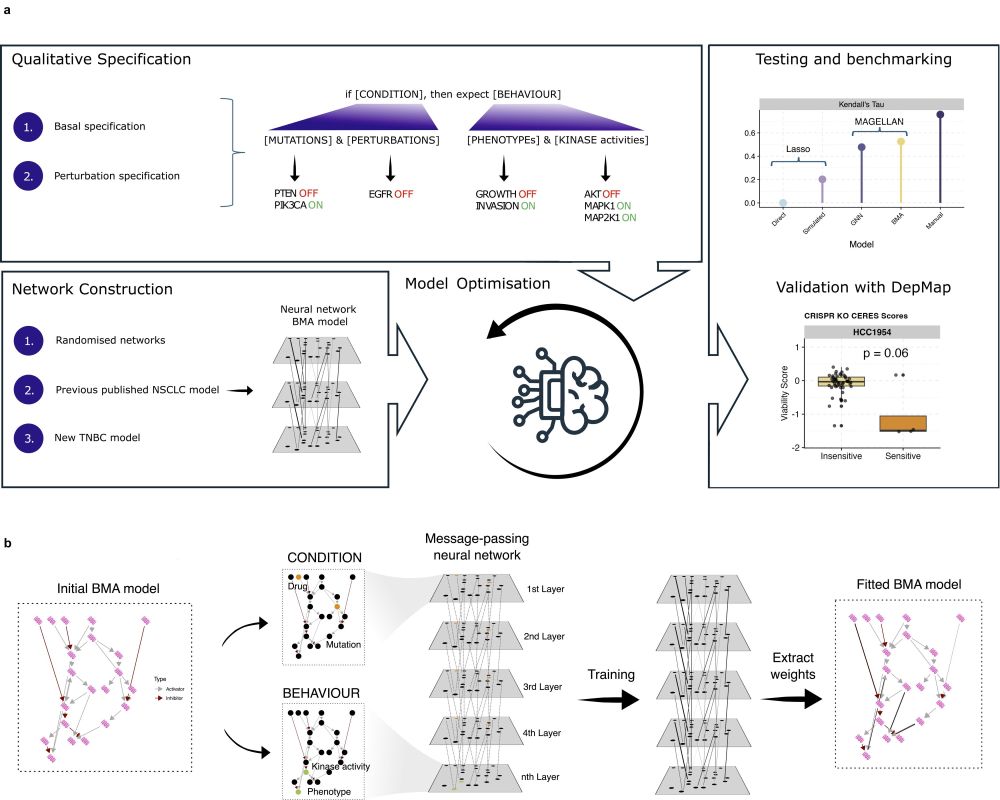
Outline of the workflow and basic fitting of a computational model to the literature derived
specification using MAGELLAN
Excited to share our new preprint showcasing a method to infer transparent, mechanistic network models of biological pathways using graph neural networks! www.biorxiv.org/content/10.1...
26.05.2025 21:49 — 👍 9 🔁 2 💬 1 📌 0
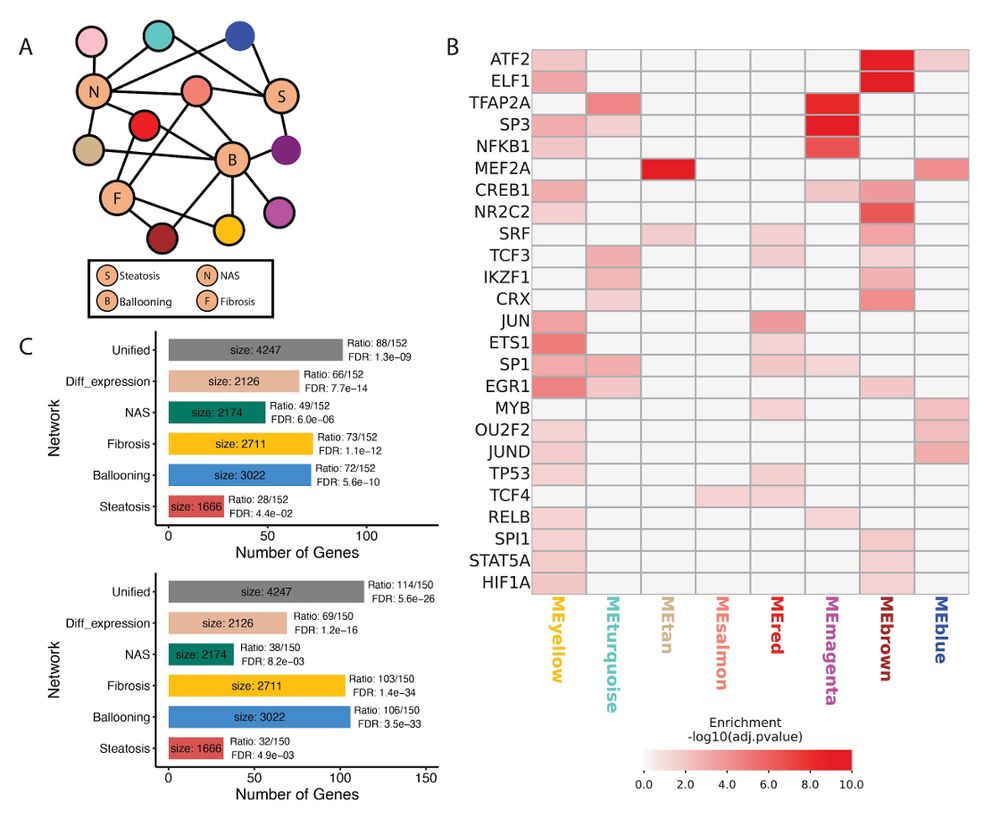
We then used our published method from @charliegbarker.bsky.social (Barker et al, Genome Research, 2022) to create a reference MASLD network by associating gene co-expression modules with histology scores, identifying key transcription factors regulating them and applying network reconstruction.
19.01.2025 11:18 — 👍 1 🔁 2 💬 1 📌 0

Excited to be working on gradient routing as part of Supervised Program for Alignment Research with Alex Cloud, Ariana Azarbal, Cailley Factor & Jorio Cocola. We hope to build safer AI by controlling where and how neural networks learn. See here for more: www.lesswrong.com/posts/tAnHM3...
28.03.2025 17:46 — 👍 5 🔁 0 💬 0 📌 0
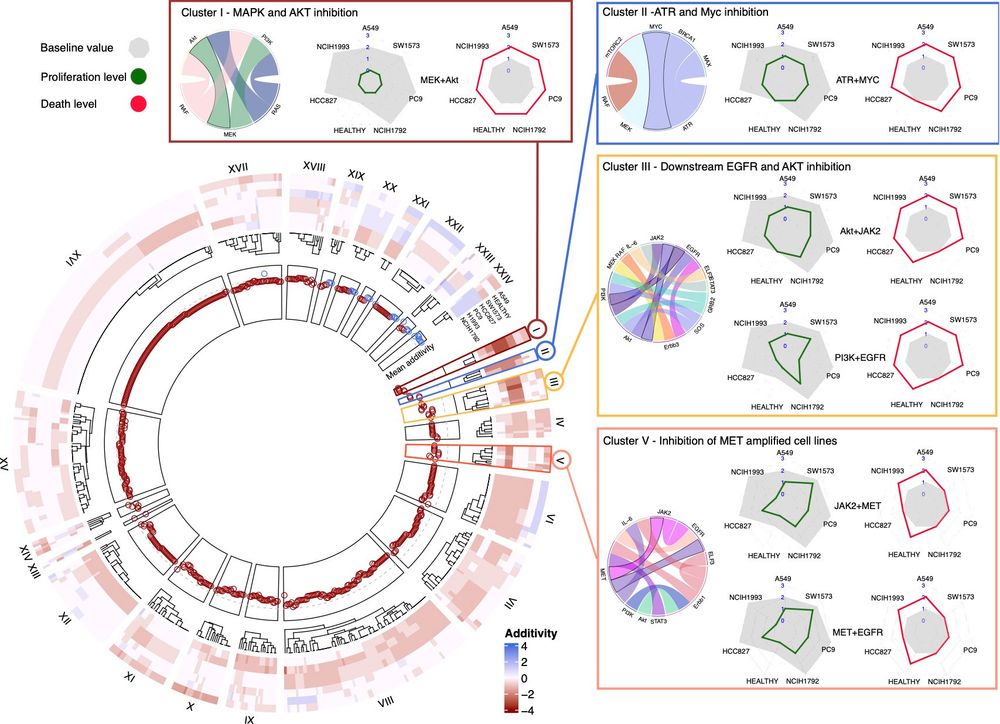
🚨 New preprint!🚨 Tackling non-small cell lung cancer (NSCLC) adenocarcinoma with in silico models:
🌟 Models tumor pathways (EGFR, AKT, JAK/STAT, WNT)
🌟 personalized drug & radiotherapy combos
🌟 Identifies a signature for patient stratification
13.01.2025 11:04 — 👍 6 🔁 2 💬 1 📌 0

Special thanks to my co-first authors @charliegbarker.bsky.social and Ashley Nicholls for their incredible work on this project, and to the Jasmin Fisher lab and all our collaborators!
12.01.2025 17:29 — 👍 1 🔁 0 💬 0 📌 0

By predicting the effect of combining targeted therapies with radiotherapy, as well as over 10,000 combinations of drugs and driver mutations, we find promising strategies to improve treatment outcomes, and identify a 19-gene signature that could help predict response to radiotherapy.
12.01.2025 17:29 — 👍 2 🔁 0 💬 1 📌 0

Excited to share our new preprint exploring the potential to personalise treatment for non-small cell lung cancer (NSCLC) using in silico modeling: www.biorxiv.org/content/10.1...
12.01.2025 17:29 — 👍 4 🔁 0 💬 1 📌 0

Huge thanks to everyone who made the work above possible, especially the @cambiochem.bsky.social , @ucllifesciences.bsky.social, Joseph Bloom, Jasmin Fisher and the Fisher Lab (www.ucl.ac.uk/cancer/resea...)
12.01.2025 15:19 — 👍 1 🔁 0 💬 0 📌 0

Applying my experience in biological networks to LLMs, I find a promising method for extracting interpretable features, Sparse AutoEncoders, is more complex than expected. SAE latents are not always independent, forming clusters mapping interpretable subspaces. (www.lesswrong.com/posts/WNoqEi...)
12.01.2025 15:19 — 👍 2 🔁 0 💬 1 📌 0
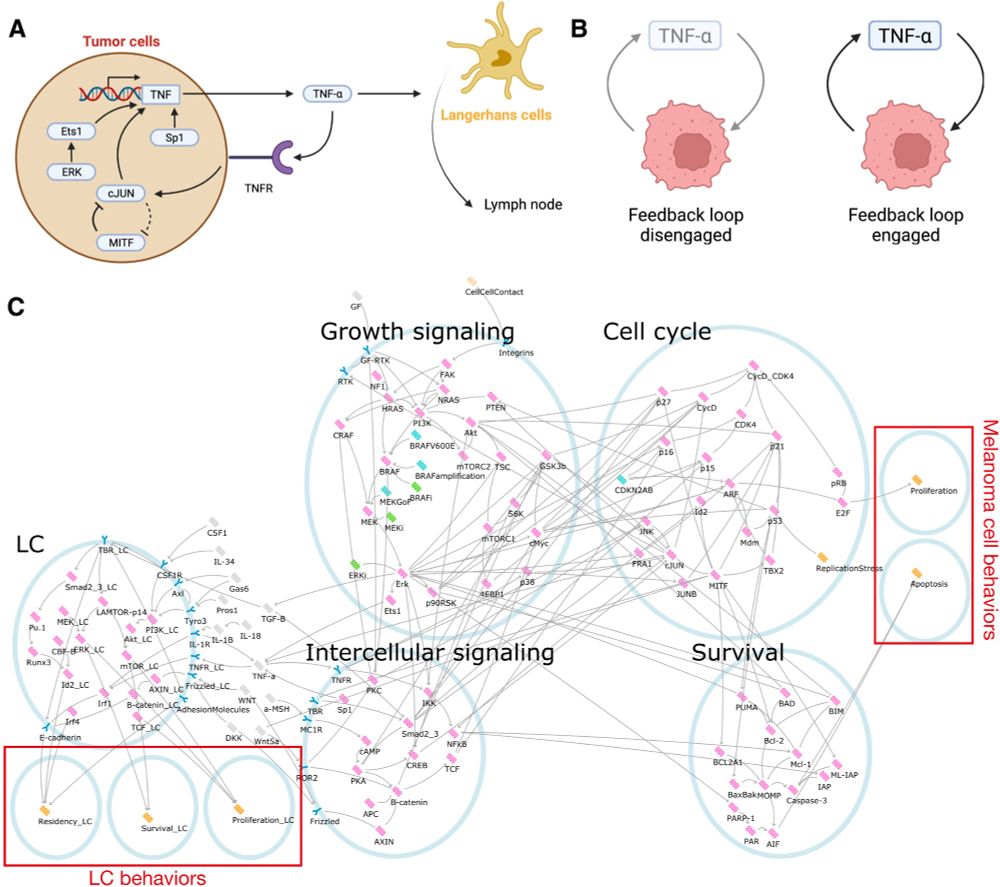
Melanomas often escape immune surveillance. Rowan Howell led work to discover why using network modelling, finding that this rests on a TNF-alpha feedback loop that prevents Langerhans cell migration to the lymph nodes. (www.science.org/doi/full/10....)
12.01.2025 15:19 — 👍 1 🔁 0 💬 1 📌 0

The behaviour of myeloproliferative neoplasms depends not only on what mutations they have, but the order in which they acquired them. Laure Talarmain led research to work out why, finding that HOXA9 acts like a switch that remembers which mutation occurred first. (www.nature.com/articles/s41...)
12.01.2025 15:19 — 👍 1 🔁 0 💬 1 📌 0

During the pandemic, we rapidly pivoted our methods for finding cancer combination therapy to COVID-19, identifying treatments most likely to be effective for different stages of the disease, as well as remaining useful even with likely resistance mutations. (www.nature.com/articles/s41...)
12.01.2025 15:19 — 👍 1 🔁 0 💬 1 📌 0

Professor Jasmin Fisher and I reviewed the exciting progress in using executable computational models to understand cancer complexity. These offer the opportunity to integrate diverse data to predict response and resistance, paving the way for personalized treatment (www.nature.com/articles/s41...)
12.01.2025 15:19 — 👍 1 🔁 0 💬 1 📌 0

We investigated cooperation between cancer cells in silico and in vivo, showing how high-Myc expressing cells drive tumour growth but become dependent on WNT-expressing cells, and found combinations of drugs that best disrupted this cooperation. (www.pnas.org/doi/abs/10.1...)
12.01.2025 15:19 — 👍 1 🔁 0 💬 1 📌 0

Based on work by Garg et al. (academic.oup.com/bioinformati...), we took advantage of binary decision diagrams to efficiently search future and past possible states of genetic regulatory to understand how past mutations restrict future evolution. (api.repository.cam.ac.uk/server/api/c...)
12.01.2025 15:19 — 👍 1 🔁 0 💬 1 📌 0

To get things started on Bluesky, here is a thread of the research I have been part of so far! For details on all of these and more, please see my website: mclarke1991.github.io
12.01.2025 15:19 — 👍 5 🔁 0 💬 1 📌 0
I got a bsky so that i could repost this!! Full publication to follow shortly but, would appreciate your feedback on the methods and results! 🔭🧪
10.12.2024 14:04 — 👍 9 🔁 3 💬 0 📌 0
Of aggregation, rare cancer biology, and organoids
First gen / Prof @ CU Anschutz
Co-President: @TheSFPM.bsky.social
Website & papers: https://linktr.ee/soragnilab
Twitter archive: @twitter.soragnilab.com
Here in a personal capacity.
Author, THE DEATH OF CONSENSUS (The Times Politics Book of 2022)
GHOSTS OF IRON MOUNTAIN ("riveting" - The New Yorker)
+ https://tinyurl.com/POWER-FAILURE (Future Governance Forum)
Last on Radio 4: Start the Week: https://www.bbc.co.uk/programmes/m002q2qm
Nukes at UNIDIR. Views own, RT/❤️ ≠ endorsement
Professor of European Union Law and Jean Monnet Chair, Ghent European Law Institute (GELI), UGent; Visiting Professor College of Europe
https://www.ugent.be/re/epir/en/researchgroups/european-law/department/staff/petervanelsuwege
Global energy think tank accelerating the clean energy transition with data and policy #CleanPower info@ember-energy.org
https://ember-energy.org/
Host of Local News International
Subscribe for free: http://LNI.media
Support us: http://LNI.media/membership
Day job: climate change, heat pumps, energy at Nesta
Other stuff: low-fi economics on growth, cities & economic geography, general UK policy, occasional basic charts
Bristol, he/him, lots of parenting / caring.
Personal account.
Japanese/Chinese ハーフ living in the USA
大田区🇯🇵 X 辽宁🇨🇳 X SF Bay Area🇺🇸
Has a Japanese soul, an American brain, and a Chinese stomach.
Born in Japan, roots for Japan in the Olympics🇯🇵🎌🇯🇵
Somewhat of an Japan Knower.
Loves history. Radical Qing monarchist?
ai enjoyer, human flourishing, cat ✝️
speechmap.ai
mindmeldai.substack.com
xlr8harder.substack.com
Open source #bioinformatics at Sungkyunkwan University 🇰🇷 | former Steinegger Lab @ SNU, Söding Lab @ MPI-NAT | http://mstdn.science/@milotmirdita
Developing data intensive computational methods • PI @ Seoul National University 🇰🇷 • #FirstGen • he/him • Hauptschüler
Computational Cancer Genomics Group
Senior researcher at @openmeasures.bsky.social
Co-host of "Posting Through It," a weekly podcast: https://www.patreon.com/PostingThroughIt
𝐓𝐡𝐞 𝐞𝐲𝐞 𝐨𝐟 𝐭𝐡𝐞 𝐰𝐨𝐫𝐥𝐝.
🔎 Keep exploring → civixplorer.com
🗺️ Get your 3D relief map (𝟭𝟬% 𝗼𝗳𝗳) → www.3dmap.cc/coupon_civixplorer
Labour MP for Nottingham East • Nottingham born and bred • she/her 🌹🏹🏳️🌈
Updates from the Nancy Keller Lab at the University of Wisconsin-Madison
Fungal genetics, genomics, and secondary metabolism. 🍄 Managed by grad students
Website: https://sites.google.com/view/kellerlabuw/keller-lab-home?authuser=0
Russia & Economics
All views private
German Institute for International and Security Affairs (SWP Berlin)
Picture: Tamek Kowalski
Labour MP for Bethnal Green and Stepney.
Computational protein engineering & synthetic biochemistry at Takeda
Opinions my own
https://linktr.ee/ddelalamo
Associate Political Editor @NewStatesman. Devout classicist, "indulgent editrix", at one point the only Ancient Greek teacher in South Korea


















