Notably, we have successfully used this tool to identify a previously unknown transcription factor for negative regulation of gliotoxin and found the bZIP protein AtfA as required for fungal responses to lipo-chitooligosaccharides (LCOs) microbial signals.
08.01.2026 19:20 — 👍 1 🔁 1 💬 0 📌 0

A network-based model of Aspergillus fumigatus elucidates regulators of development and defensive natural products of an opportunistic pathogen
Abstract. Aspergillus fumigatus is a notorious pathogenic fungus responsible for various harmful, sometimes lethal, diseases known as aspergilloses. Unders
We are excited to announce the publication of GRAsp, a gene regulatory network prediction tool for A. fumigatus in collaboration with @sroyyors.bsky.social and @jeanmichelane.bsky.social in Nucleic Acid Reports!
This tool can be found on our lab website or at grasp.wid.wisc.edu.
08.01.2026 19:11 — 👍 5 🔁 2 💬 1 📌 0

network-based model of Aspergillus fumigatus elucidates regulators of development and defensive natural products of an opportunistic pathogen
Aspergillus fumigatus is a notorious pathogenic fungus responsible for various harmful, sometimes lethal, diseases known as aspergilloses. Understanding the gene regulatory networks that specify the expression programs underlying this fungus’ diverse phenotypes can shed mechanistic insight into its growth, development, and determinants of pathogenicity. We used eighteen publicly available RNA-seq datasets of Aspergillus fumigatus to construct a comprehensive gene regulatory network resource. Our resource, named GRAsp (Gene Regulation of Aspergillus fumigatus), was able to recapitulate known regulatory pathways such as response to hypoxia, iron and zinc homeostasis, and secondary metabolite synthesis. Further, GRAsp was experimentally validated in two cases: one in which GRAsp accurately identified an uncharacterized transcription factor negatively regulating the production of the virulence factor gliotoxin and another where GRAsp revealed the bZip protein, AtfA, as required for fungal responses to microbial signals known as lipo-chitooligosaccharides. Our work showcases the strength of using network-based approaches to generate new hypotheses about regulatory relationships in Aspergillus fumigatus. We also unveil an online, user-friendly version of GRAsp available to the Aspergillus research community.
Latest paper from our lab in collaboration with @sroyyors.bsky.social and Nancy Keller.
Most notably for me, we describe here the first fungal gene required for responses to LCOs (Nod factors) in the fungus Aspergillus fumigatus. More to come on this topic later!
08.01.2026 19:06 — 👍 6 🔁 3 💬 0 📌 0
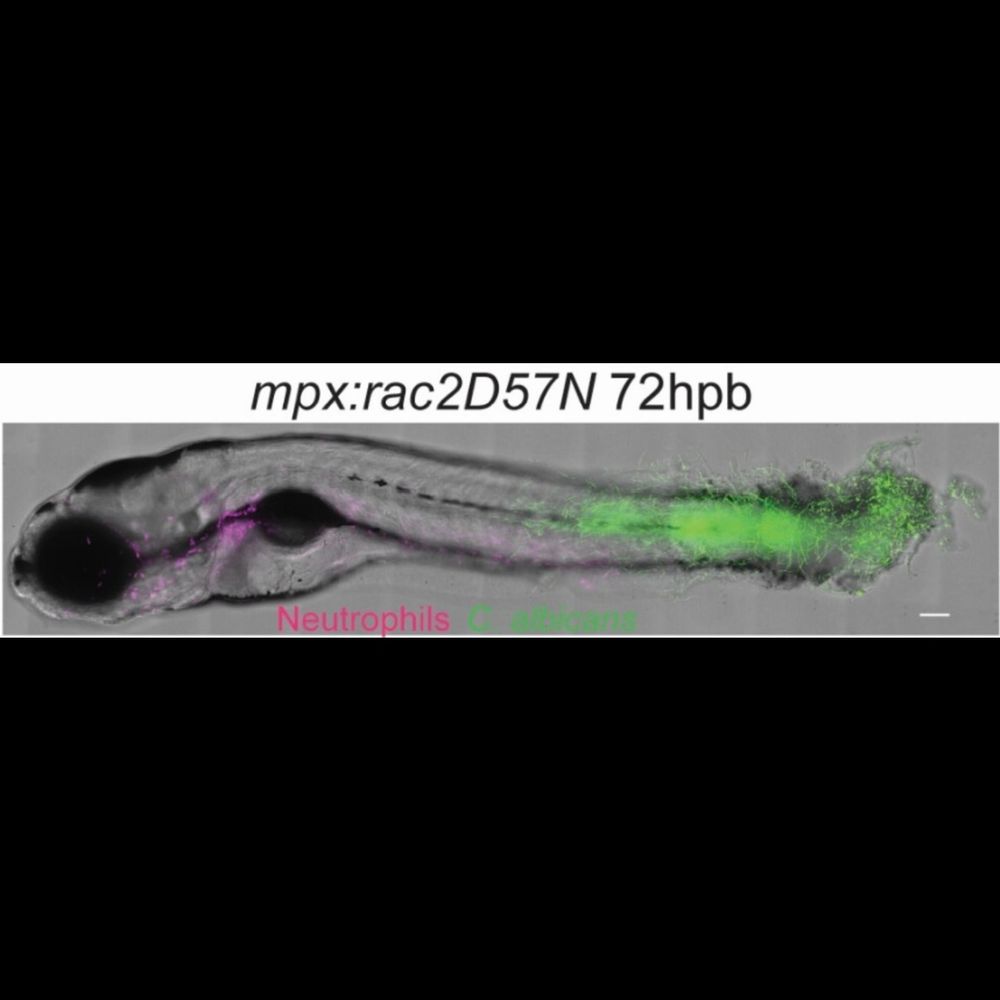
Importantly, we also show the strength of the zebrafish larval model to study virulence factors across fungal species.
16.04.2025 18:46 — 👍 0 🔁 0 💬 0 📌 0
Microbiology Society: A world in which the science of #microbiology provides maximum benefit to society | microbiologysociety.org
lnk.bio/microbiosoc
Regulatory genomics, machine learning, networks, systems biology, evolution
Professor of Microbiome Ecology at Institute of Biology, Leiden University
Bacillus subtilis, biofilms, plant microbiome, bacteria-fungi interactions and circadian clock within @microclockerc.bsky.social #ERCSyG
https://linktr.ee/atkovacs
Associate Professor Tufts University
Ecology/evolution of microbes in food systems. Constant gardener.
Bioinformatician working in microbial genomics, hiking fan and reading fanatic | she/her | opinions are my own & may not reflect the views of my employer or funding agency
Fungi >>>
annehatmaker.com
microbiologist. entomologist. immunologist. Lover of science, especially at its intersections. Asst Prof of Biology at Johns Hopkins. @nabroderick elsewhere.
Bacterial geneticist focused on microbe-animal communication. Professor UW-Madison. Messages are my personal views.
PhD candidate in the Heitman lab @dukemedschool studying genetics of antimicrobial resistance in fungal pathogens | @dartmouthartsci alum | runner, skier, outdoor enthusiast
Interested in microbial ecology & evolution. Views are only my own. (he/him)
🎓: https://scholar.google.com/citations?hl=en&user=OBPpZq4AAAAJ&view_op=list_works&sortby=pubdate
👨💻: https://github.com/raufs
Aspergillus Genomes Research Policy Committee
Organisers of annual Asperfest meeting
***Asperfest21 in Dublin March 2nd 2025!***
www.aspgrpc.com
The Mycological Society of America is a professional society for the study of fungi and fungal-like organisms & publishes the journal #Mycologia. #MSAfungi26
Graduate student | Microbiology Doctoral Training Program | University of Wisconsin-Madison | Jason Peters lab | Pathogens | Antibiotic Resistance | CRISPR
The Canadian Fungal Network brings together diverse groups of scientists whose interests span the breadth of fungal research in Canada and abroad.
https://fungalresearch.weebly.com/
CanFunNet25 conference site: https://canfunnet.conferences.uwo.ca/
Junior group leader at the University of Jena (@uni-jena.de)
CIFAR Azrieli Global Scholar
Fungal pathoecology, antimicrobial resistance, and genome stuff.
Always up for an adventure or bike ride.
https://barber-lab.com
Microbiome gen(om|et)ics, secondary metabolism, comp bio, antibiotic/drug disco, evolution, & hockey. he/him.
Gut microbiome, bacterial pathogens, bacteriophages.
Assistant Professor, Depts. Medicine and Medical Microbiology & Immunology, University of Wisconsin-Madison. Views expressed = mine. hrycklab.medicine.wisc.edu
We are an interdisciplinary institute at the University of Wisconsin–Madison exploring simplicity in complex systems at the intersection of digital and natural worlds.
Official Bluesky account for the UW Madison MDTP
Science and research news from the University of Wisconsin-Madison. https://news.wisc.edu
Professor at the University of Wisconsin - Madison. Researcher on plant-microbe symbioses. Father of 5. Loves hiking, camping, archery, and coffee. Views are my own.