Home - Sakaue Lab @ Genome Sciences, UW
Lab website for Dr. Saori Sakaue
I’ll be at #ashg2025 ! Please let me know if you want to chat with me about research, my lab at UW saorisakaue.github.io etc etc! I’ll also be standing at the poster session on Friday 5075F at 2:30-4p if you can swing by to chat👋
14.10.2025 17:22 — 👍 1 🔁 0 💬 0 📌 0

New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...
26.09.2025 18:06 — 👍 53 🔁 20 💬 3 📌 2
The project was super fun and unexpected scientific journey, expanding my curiosity outside of the nucleus and DNA in genetics studies😀 I really appreciate warm mentorship from
@soumya-boston.bsky.social and invaluable inputs from genetics community in Boston! Hope you enjoy reading the preprint😉
27.02.2025 02:29 — 👍 3 🔁 0 💬 0 📌 0
Back to the original problem🔍 We've found that >50% of colocalization of causal variants between eQTL and neuropsychiatric disorders were specific to either nuclear or cytosolic eQTL! Subcellular localization of RNA and eQTL matters in identifying disease GWAS mechanisms (11/n
27.02.2025 02:28 — 👍 0 🔁 0 💬 1 📌 0

We've conventionally assumed that multiple causal variants in eQTL or GWAS is typically LD independent and working on different biological mechanisms (eg. enhancer AND promoter), but in our cases they can be LD *dependent* and work in concert to affect the same mechanism of RNA stability (10/n
27.02.2025 02:28 — 👍 1 🔁 0 💬 2 📌 0

Such examples with different causal variants between nucleus (enhancer) and cytosol (transcript) showed that sometimes many variants in complete linkage in cytosolic eQTL create a risk haplotype in the transcript, possibly suggesting a novel concept in multiple-causal-variant fine-mapping.(9/n
27.02.2025 02:27 — 👍 0 🔁 0 💬 1 📌 0
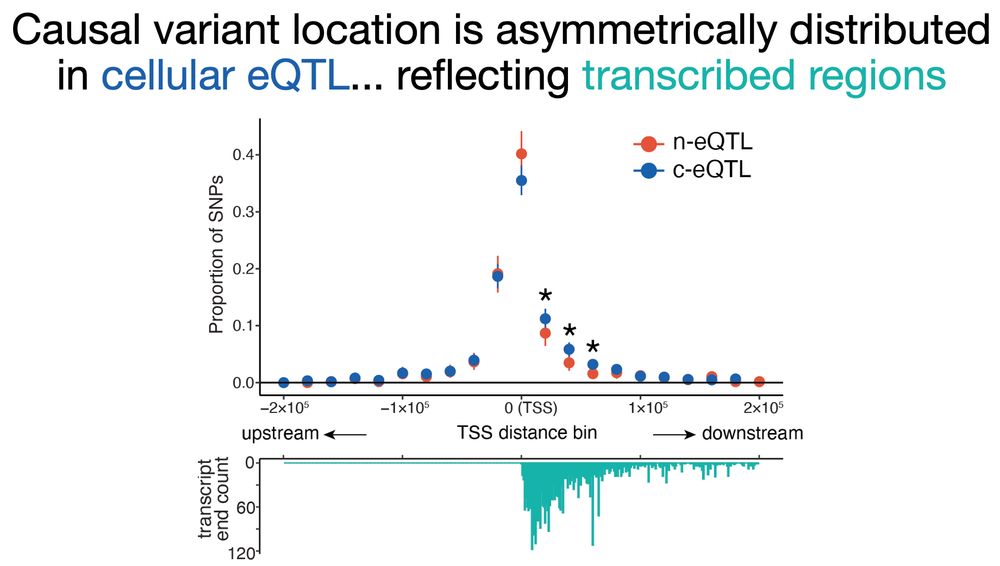
.. which makes cytosolic eQTL variants more asymmetric, downstream-skewed relative to the TSS as they localize within transcribed regions where RNA chemical modification can happen and affect RNA stability in the cytosol. (8/n
27.02.2025 02:26 — 👍 0 🔁 0 💬 1 📌 0

Surprisingly, 33% of eGenes had distinct causal variants between nucleus and cytosol for the same gene!
Nuclear early RNA was preferentially regulated by distal enhancers at the DNA transcription level, while cytosolic late RNA was regulated by variants within transcripts.. (7/n
27.02.2025 02:26 — 👍 0 🔁 0 💬 1 📌 0

More specifically, we asked if nuclear and cellular eQTL share the same causal variant (1), and if they are different, which genomic annotation(s) has preferential localization of causal variants for nuclear or cellular eQTL (2)? (6/n
27.02.2025 02:25 — 👍 0 🔁 0 💬 1 📌 0

We analyzed both nuclear and cellular (mostly cytosolic) RNA compartments and associated their RNA abundance with genotype in the brain and the kidney to achieve this goal! (5/n
27.02.2025 02:24 — 👍 0 🔁 0 💬 1 📌 0

So we asked distinct genetic regulatory mechanisms across entire RNA lifecycle by comparing eQTL between early RNA in the nucleus and late post-transcriptionally modified RNA in the cytosol. These molecular understanding will help us understand disease alleles precisely (4/n
27.02.2025 02:24 — 👍 0 🔁 0 💬 1 📌 0

But the DNA transcription into RNA is just the very first step among long journey of RNA lifecycle. RNA undergoes many processing, first in the nucleus e.g. splicing and polyA, then in the cytosol e.g. chemical modifications and degradation essential for mature RNA abundance (3/n
27.02.2025 02:23 — 👍 0 🔁 0 💬 1 📌 0

Problem: eQTL reveals disease alleles' function on gene expression, while it's been so puzzling🧐 that most #GWAS alleles do not colocalize with #eQTL. The traditional wisdom in the field is that eQTL regulate DNA transcription in the nucleus by altering regulatory sequences (2/n
27.02.2025 02:22 — 👍 0 🔁 0 💬 1 📌 0
📣Excited to share my last postdoc paper with
@soumya-boston.bsky.social on eQTL mechanisms depending on where the RNA is in the cell! @broadinstitute.org @harvardmed.bsky.social
TL;DR:Early RNA eQTL variants in the nucleus and late RNA eQTL variants in the cytosol have distinct molecular mechanism🧵
27.02.2025 02:21 — 👍 71 🔁 22 💬 2 📌 1
Nobuhiko (Nobu) Hamazaki, Ph.D. Assistant prof. @University of Washington (UW). Stem cell engineering, germ cell and embryonic development.
PhD Student @ UW Genome Sciences
The Molecular and Cellular Biology (MCB) program is an interdisciplinary graduate program jointly offered by the University of Washington and the Fred Hutchinson Cancer Center. https://mcb-seattle.edu/
Working to realize the promise of human genomics to better predict, prevent, and treat cancer. For more info: https://labs.dana-farber.org/collins-genomics/
Based in the Department of Oncology at the MRC WIMM, University of Oxford | We study the genetics of immunity with a focus on the determinants of response to cancer immunotherapy.
Genetics, immunology, oncology, science.
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Assistant prof at UCLA using the Bayes for the genomes https://pimentellab.com
postdoc at UW Genome Sciences with @wsdewitt.github.io | previous: PhD student with @matsen.bsky.social at Fred Hutch |
♡ computational biology, immunology
There is a crack in everything, that’s how the light gets in.
Scientist, Computational biologist. Asst. Prof. Icahn School of Medicine at Mount Sinai 🧬. Alum. Harvard Medical School 🇺🇸, EPFL🇨🇭, University of Tehran 🇮🇷
Your complete source for all the life science news and events in the Seattle area. Brought to you by the Science in the City program @stemcell.com.
Scientist at UW Genome Sciences and the Seattle Hub for Synthetic Biology.
http://pinglay-lab.com/
synBio/genomics/soccer/heavy metal/food
Graduate student in the Dunham Lab within the Department of Genome Sciences at The University of Washington
Assistant Professor @ UCLA Pathology, Computational Medicine, Biostatistics, and Genetics
WashU Immunology
Graduate student interested in T cell exhaustion.
https://egawalab.wustl.edu
Instructor at Boston Children’s Hospital, Harvard Medical School | Broad Institute.
#bioinformatics #genomics
Scientist at DKFZ and EMBL in Heidelberg, loving stats, genomics and genetics. @OliverStegle@genomic.social. For group news see @steglelab.bluesky.social
Principal Investigator at Boston Children's Hospital | Statistical Genetics
Scientist!
Systems Analyst and Computational Biologist, PhD.
https://katiaplopes.github.io/
Associate Professor
DFCI & HMS











