https://www.pinglay-lab.com/
if you are excited about these areas, come join our lab
@uwgenome.bsky.social and the Seattle Hub for Synthetic Biology! We are hiring at all levels.
t.co/wAehoBsJ5F
23.01.2026 19:12 — 👍 1 🔁 0 💬 0 📌 0

We also discuss current bottlenecks for mammalian genome writing (mainly throughput and size), mechanisms to address them, and future directions for the technology. We think it is particularly poised to revolutionize generating training data for genomic AI models.
23.01.2026 19:12 — 👍 1 🔁 0 💬 1 📌 0

Engineering mammalian cells with large synthetic DNA payloads promises to be highly enabling for understanding mammalian gene regulation, dissecting GWAS haplotypes, engineering better animal models for human disease, and encoding multi-gene functions for cell therapy and biotech
23.01.2026 19:12 — 👍 0 🔁 0 💬 1 📌 0

Please 🔁! #MASSIV will be held in Vancouver from January 19-22, 2026. 4-day meeting on #synbio x #tissueengineering.
Some trainee-focused events. >50 talks (10 from abstracts), posters, panel & publishing insights sessions.
www.massivconference.com
Abstract deadline: Nov 14
25.10.2025 16:21 — 👍 4 🔁 7 💬 1 📌 1

Please RP! A 4-day synbio and tissue engineering conference #MASSIV to transform biology and medicine together.
Couldn't list all amazing speakers, so please check out this 👉https://www.massivconference.com/
Poster due is Nov 7, but will be extended.
With @stemcellnetwork.ca and JST.
18.10.2025 17:50 — 👍 4 🔁 6 💬 0 📌 0

New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...
26.09.2025 18:06 — 👍 54 🔁 20 💬 3 📌 2

Jobs
We are working to solve the biggest mysteries in bioscience.
We are hiring a scientist to lead our 'Build' team at the Seattle Hub for Synthetic Biology. @alleninstitute.org
Job posting below:
alleninstitute.org/careers/jobs...
Please reach out if you have any questions!
13.08.2025 19:03 — 👍 7 🔁 6 💬 0 📌 0

Pinglay Lab
A big thank you to all co-authors - looking forward to seeing where we can take this approach.
If you are interested in joining us on this effort, check out our website: www.pinglay-lab.com
14.07.2025 22:41 — 👍 1 🔁 0 💬 1 📌 0
Finally, we demonstrate that data resulting from SGE is compatible with training predictive machine learning models.
We are very excited about using SGE to generate the synthetic data needed to train the next generation of models for biological design.
14.07.2025 22:41 — 👍 0 🔁 0 💬 1 📌 0

SGE is generalizable across cell types. We engineer T-cells (Jurkat) to grow without valine.
We believe a similar strategy could help create more resilient T-cells for therapy, capable of surviving and functioning in the metabolically depleted environments of tumors. Hopefully more here soon!
14.07.2025 22:39 — 👍 1 🔁 0 💬 1 📌 0

Pinglay Lab
A big thank you to all co-authors - looking forward to seeing where we can take this approach.
If you are interested in joining us on this effort, check out our website: www.pinglay-lab.com
14.07.2025 22:36 — 👍 0 🔁 0 💬 1 📌 0
Finally, we demonstrate that data resulting from SGE is compatible with training predictive machine learning models.
We are very excited about using SGE to generate the synthetic data needed to train the next generation of models for biological design.
14.07.2025 22:36 — 👍 0 🔁 0 💬 1 📌 0

We then used SGE to engineer CHO cells to grow without isoleucine, a feat we could not achieve via rational design and delivery of entire synthetic pathways.
Again, mitochondrial localization was favored, with individual clones reflecting ~40-50kb of integrated DNA!
14.07.2025 22:36 — 👍 1 🔁 0 💬 1 📌 0

Using SGE, we screened millions of pathway combinations in a single experiment to engineer CHO cells that grew at WT rate (~1.1 day/doubling) in valine-free medium.
Intriguingly, the best clones all employed mitochondrial localization of pathway components, not cytoplasm as in our prev. design.
14.07.2025 22:36 — 👍 1 🔁 0 💬 1 📌 0

Resurrecting essential amino acid biosynthesis in mammalian cells
Mammalian cells were engineered to synthesize valine, a metabolic capacity that had been lost from the lineage of higher eukaryotes for >500 million years.
In collaboration with Harris Wang’s lab, we previously engineered cells to grow without valine by importing 4 genes from E.coli.
However, the cells grew 4x slower than normal - and we could not extend this strategy to enable any other amino acid prototrophies.
doi.org/10.7554/eLif...
14.07.2025 22:36 — 👍 1 🔁 0 💬 1 📌 0

As a test case, we used SGE to engineer essential amino acid prototrophy in mammalian cells, a behavior last seen over 500 million years ago.
Unlike E. coli, which can make all 20 proetinogenic amino acids, mammals lack the pathways for 9 “essential” ones and must obtain them through the diet.
14.07.2025 22:36 — 👍 1 🔁 0 💬 1 📌 0

In SGE, we clone and deliver a TU library at high MOI so that each cell gets a random mix, assembling a unique synthetic metabolic pathway per cell. Cells with the desired phenotype (e.g., survival or fluorescence) are selected, and TU barcodes are sequenced to identify functional combinations.
14.07.2025 22:36 — 👍 0 🔁 0 💬 1 📌 0
To address this, we developed Shotgun Genetic Engineering (SGE), which leverages the fact that building and delivering many small, barcoded transcription units - each with a gene, promoter and localization signal - is exponentially easier than delivering a single large construct to a mammalian cell.
14.07.2025 22:36 — 👍 0 🔁 0 💬 1 📌 0

Mammalian metabolic engineering is key to advancing bioproduction, cell therapy, and rejuvenation.
But as pathway complexity grows, so does the combinatorial design space! However, delivering large DNA constructs to mammalian cells is inefficient, making large unbiased screens intractable.
14.07.2025 22:36 — 👍 0 🔁 0 💬 1 📌 0

A shotgun approach for highly multiplexed mammalian metabolic engineering
Mammalian metabolic engineering is critical to advancing basic biology, bioproduction, and cell therapy. However, as pathway complexity increases, so does the size of both the combinatorial design spa...
Excited to share our work with @julietrolle.bsky.social & Jef Boeke.
We developed a 'shotgun' method to screen millions of synthetic metabolic pathways to enable mammalian cells to grow without two essential nutrients for the first time in >500 million years!
doi.org/10.1101/2025...
Thread...
14.07.2025 22:36 — 👍 23 🔁 10 💬 1 📌 0

Range extender mediates long-distance enhancer activity - Nature
The REX element is associated with long-range enhancer–promoter interactions.
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
02.07.2025 16:17 — 👍 186 🔁 90 💬 10 📌 9
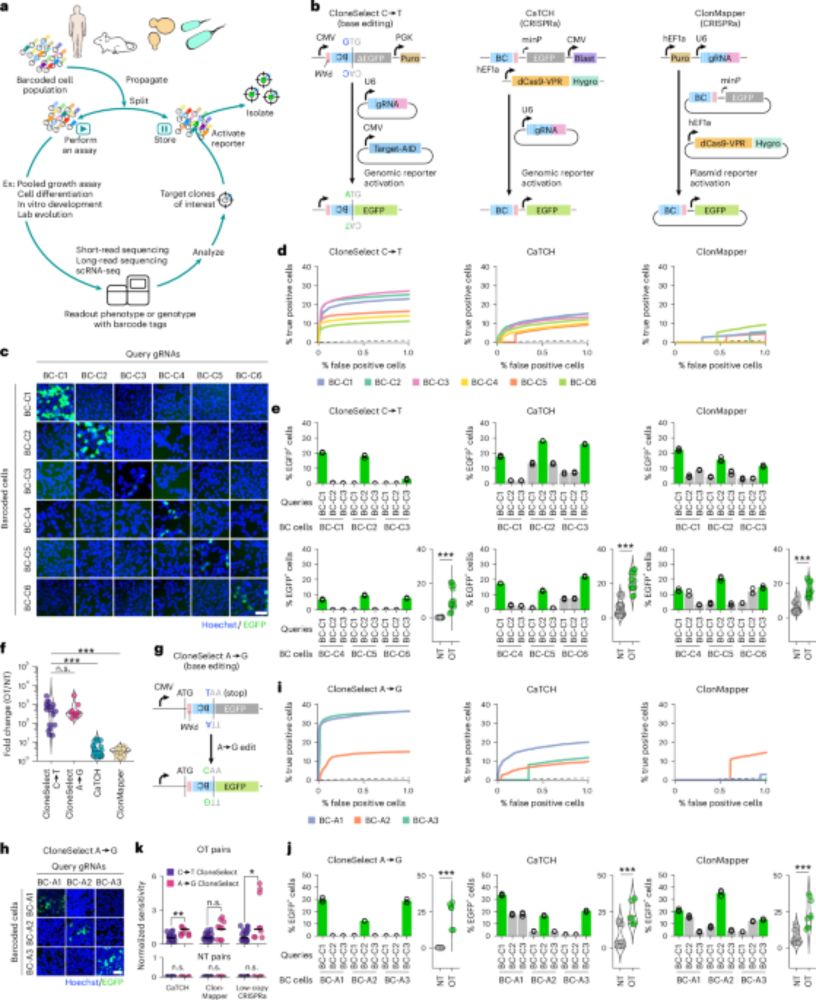
A multi-kingdom genetic barcoding system for precise clone isolation - Nature Biotechnology
A barcoded CRISPR base editing system isolates target clones from complex mammalian, yeast and bacterial populations.
CloneSelect published in Nature Biotechnology
@natbiotech.nature.com. This retrospective clone isolation method using CRISPR base editors is a powerful tool in broad biology. A history of Soh in the Yachie lab. www.nature.com/articles/s41...
21.05.2025 13:56 — 👍 29 🔁 13 💬 3 📌 0
Such a cool story Maximus! Congrats
25.04.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0
Let’s gooo! Congrats Dave and team - glad to see this out.
11.04.2025 14:58 — 👍 2 🔁 0 💬 0 📌 0
Studying The Dark Matter of the Genome - everything noncoding 🧬
A collaborative scientific project at NYU Langone.
Account managed by Ran Brosh.
https://orcid.org/0000-0003-3471-4637
Our research focuses on the impact of compressive stress on the living, and the interplay with intracellular crowding, from an engineering, physical and biological point of view. Check out our website https://delarue-research.org/ for more information!
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan
Previously IMP Vienna, Stanford Genetics, UW CSE.
Computational biologist interested in deciphering the genomic regulatory code at vib.ai
Neuroscience postdoc at NYU Grossman School of Medicine researching glial cell contributions to neurodegeneration. https://www.pryprk.com/
MD/PhD Univ Wash, PhD candidate in Jay Shendure lab | Yale '18 | NRSA Fellow | genomics & precision medicine | she/her | views & opinions are my own 🧬 💫
Postdoc - Duboule Lab - @college-de-france.fr @cirbcdf.bsky.social
Formerly PhD student - Furlong Lab - @EMBL.org
Interested in gene expression and genome topology regulation during embryogenesis
From 🇮🇹 , then Heidelberg 🇩🇪, now Paris 🇫🇷
This is the BlyeSky account of the Night Science Institute - night-science.org - which champions a cultural shift in science by training researchers to embrace the creative Night Science process as an essential complement to rigorous hypothesis testing.
Engineering mammalian cells | Boeke Lab, NYU
EPIGENETIC HULK SMASH PUNY GENOME. MAKE GENOME GO. LOCATION: NOT CENTROMERE, THAT FOR SURE
Living a life of troubleshooting🫠
🐶Univ. of Washington, JSPS Overseas Postdoc fellow ←🌱Univ. of Tsukuba, Japan.
Arata Wakimoto / 脇本 新
JP/EN
#StemCells, #DevBio, #EvoDevo
Love☕ #Coffee and 👾 #Games
Assistant Prof. @uoknightcampus.bsky.social. Cofounder @synplexity.bsky.social. Gene synthesis, synbio, protein engineering, nanopores, multiplex assays. Opinions my own. www.plesalab.org
Assistant Professor at Baylor College of Medicine & Texas Children's Hospital | Human genetics and single-cell genomics | Formerly Columbia Med & Duke
We’re on a mission to end the headaches of ordering synthetic DNA and give researchers what they really want: reliable DNA, on time, exactly as ordered. #DNASynthesis 🧪
ansabio.com






















