Whats better than "publish or perperish" and "publish high or perish"?
"publish high as a first or last author or perish" !
Sometimes I feel like the people who invent these metrics have never actually done academic research.
26.10.2025 18:15 — 👍 1 🔁 0 💬 0 📌 0
Extremely important K+ and Ca2+ indicator development work in these papers.
22.10.2025 17:35 — 👍 4 🔁 4 💬 0 📌 0

🏆 First Prize – DevBio Art Contest 🖼️
“A Model Meeting” by Karla Akari Garcia
Inspired by The Quaker Meeting, this watercolor honors animal research models, from mouse to axolotl, that have advanced developmental biology 🎨 A tribute to science’s often unsung heroes. #DevBioArt
22.10.2025 20:19 — 👍 36 🔁 10 💬 3 📌 2

Ran et al. generated a retinal regeneration model and a tumor model using targeted integration in X. tropicalis (PNAS). Kagawa et al. provide a step-by-step protocol for knock-in targeted integration in X. laevis (DGD). www.xenbase.org/xenbase/doNe... #science #devbio #xenopus #frogs
21.10.2025 22:18 — 👍 5 🔁 3 💬 0 📌 0

Screenshot of the original published figure (left) and the corrected figure (right). In the left photo we see tumors in a grid of 5 columns and 6 rows. Two sets of tumors are marked by me with blue and green boxes respectively.
In the right photo the duplicated tumors are just left out.
Hey @tandfresearch.bsky.social, - you should not allow authors to address photoshopped tumors by just leaving out the photoshopped tumors. This paper was 1 year old at the time of correction. Where are the originals?
Horrible editorial decision.
#ImageForensics
20.10.2025 21:21 — 👍 52 🔁 6 💬 0 📌 1
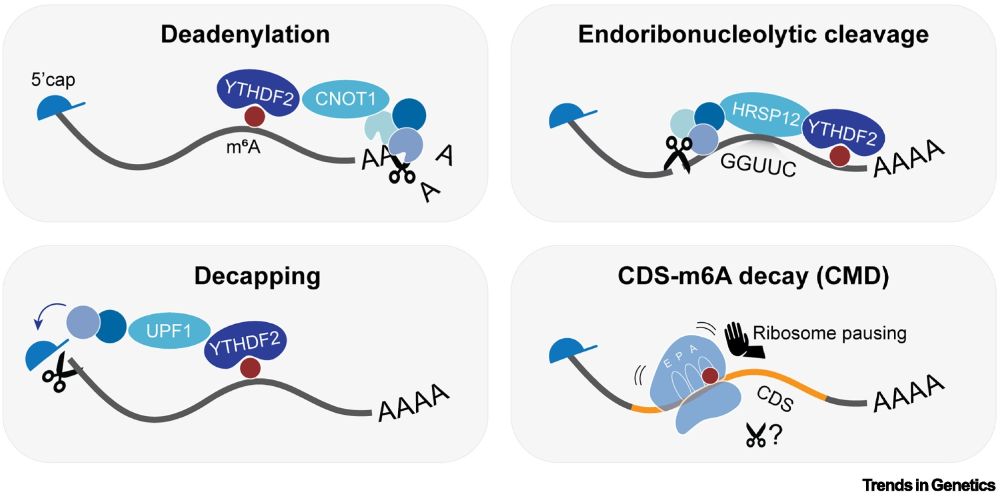
Figure 2. Overview of N6-methyladenosine (m6A) decay pathways. m6A acts as a destabilizing mark on mRNA.
"m6A in the coding sequence: linking deposition, translation and decay"
by Kathi Zarnack (@zarnack-group.bsky.social) & colleagues
"This review explores [m6A in] mRNA decay, with a focus on the newly discovered CDS–m6A decay (CMD) pathway..."
Read it here:
authors.elsevier.com/sd/article/S...
08.07.2025 12:07 — 👍 30 🔁 16 💬 0 📌 1
On #WorldMentalHealthDay, let's take a moment to pause and reflect 💚
Working in life sciences often means juggling a lot at once, and it's okay to seek support. Talking to colleagues, friends, or a professional is a strength, not a stigma.
#ItsOkayToAskForHelp #MentalHealthDay
10.10.2025 12:58 — 👍 7 🔁 1 💬 0 📌 0

Lost Science is a new NYT series of accounts from scientists who have lost their jobs or funding. You can send your story to the Times here www.nytimes.com/2025/10/08/c...
10.10.2025 08:38 — 👍 109 🔁 75 💬 8 📌 5
Don't forget the "Do you own research" fraction of "independent thinkers" who "don't just trust the main stream" and all the other people who would rather drink bleach because some person on the internet told them then consulting a medical doctor.
18.09.2025 07:56 — 👍 0 🔁 0 💬 0 📌 0
u know who are skeptics
scientists are skeptics
being anti-science is not being a skeptic
16.09.2025 23:12 — 👍 830 🔁 142 💬 12 📌 8
Quick question to the scientific hive mind: does anybody has some tips how to best transport live, adherent cells for 45 min from one lab to another?
15.09.2025 16:30 — 👍 0 🔁 0 💬 0 📌 0
Maybe, but there is this notion that, as long as ppl deliver a lot of data, everything goes. Also, most underestimate the impact of a well maintained microscope on image quality
04.09.2025 11:31 — 👍 1 🔁 0 💬 0 📌 0
TK: Authors did not reply or replied that they did not care if the paper would be retracted - they just needed it to graduate.
Retaliation: we received lots of spam after retracting papers.
We learned that paper mills can abuse peer review system. Do not rely on author-suggested reviewers.
#PRC10
03.09.2025 15:36 — 👍 14 🔁 5 💬 1 📌 1

Top: Transgenic expression of PdgfraΔK-EGFP (cyan) in nkx3.1:Gal4; UAS:NTR-mCherry (magenta) background at 56 hpf. mCherry+ EGFP+ cells are mostly restricted in the dorsal or ventral regions of the trunk. A transverse view of a 50 μm region centered on the dashed line is shown on the right, with the neural tube (nt) and notochord (n) outlined by dotted lines. Bottom: Model for how the migration of sclerotome-derived cells is regulated by chemoattraction mediated by Pdgfab/Pdgfra signaling. (A) In wild-type embryos, nkx3.1+ (cyan) sclerotome-derived cells express the receptor Pdgfra (magenta), while the medial somites express the ligand Pdgfab (orange) in response to Hh signaling. pdgfra+ sclerotome-derived cells undergo extensive migration toward both the notochord in the medial trunk and the fin fold. (B) In pdgfra−/− mutants (pdgframRFP/mRFP and pdgfraref/ref), nkx3.1+ sclerotome-derived cells fail to migrate in any direction and remain in their original location in the ventral and dorsal parts of the somites. (C) In pdgfab−/− embryos, pdgfra+ sclerotome-derived cells fail to migrate toward the notochord, although migration toward the fin fold remains unaffected. (D) In pdgfab−/− embryos with mosaic pdgfab expression, nkx3.1+ sclerotome cells can migrate toward the ectopic pdgfab-expressing cells, either medially around the notochord or ventrally toward the yolk extension. When the pdgfab source is located near or in the notochord, it can completely rescue the migration of nkx3.1+ sclerotome cells toward the notochord. These diagrams depict a 2-somite region located above the yolk extension of a zebrafish embryo at 48 hpf. nt: neural tube; n: notochord.
What guides fibroblast precursors' migration from the #sclerotome during vertebrate development? @penghuang031.bsky.social &co show that this is driven by #PDGF -mediated chemoattraction, enabling their differentiation into specialized #fibroblast subtypes @plosbiology.org 🧪 plos.io/3HTZ92Y
03.09.2025 16:42 — 👍 4 🔁 2 💬 0 📌 0
Assuming their PI cares about such things...
04.09.2025 10:05 — 👍 1 🔁 0 💬 1 📌 0
Agree. Don't know why it's so hard for some people to clean up after themselves. Sometimes I feel more scientist should learn how damn expensive this stuff is
03.09.2025 07:58 — 👍 1 🔁 0 💬 1 📌 0
Being friends with the head of our imaging facility, I learned that 9 out of 10 times blurry images at the 20x air objective are not the result of a damaged lens but of a fine crust of immersion oil on or within the objective.
02.09.2025 06:38 — 👍 3 🔁 0 💬 1 📌 0

The easiest way to get good looking fluorescence microscopy figures is to just do fine-tube them in photoshop! Silly PhD students spending years learning to create good figures. Beginners!
Huang et al. 2020 (DOI: 10.1016/j.carbpol.2020.116650) shows us how it's done!
pubpeer.com/publications...
01.09.2025 20:13 — 👍 18 🔁 3 💬 2 📌 0

Postdoc in Structural Biology of RNA processing complexes
Postdoctoral positions in structural biology of macromolecular complexes are available in the laboratory of Dr. Eva Kowalinski at the EMBL Grenoble, France. We are looking for highly motivated and amb...
Postdoc positions in structural biology of RNA processing complexes.
An ERC-funded position will dissect pre-mRNA processing pathways in the infective organism Trypanosoma brucei. An ANR-funded position will characterise large complexes in RNA modification.
Details: tinyurl.com/postdoc-RNA-...
17.07.2025 09:50 — 👍 21 🔁 19 💬 0 📌 1
Essay mills shamelessly advertising on the academia feed. CC @keithwilson.eu
21.07.2025 05:17 — 👍 21 🔁 7 💬 3 📌 2
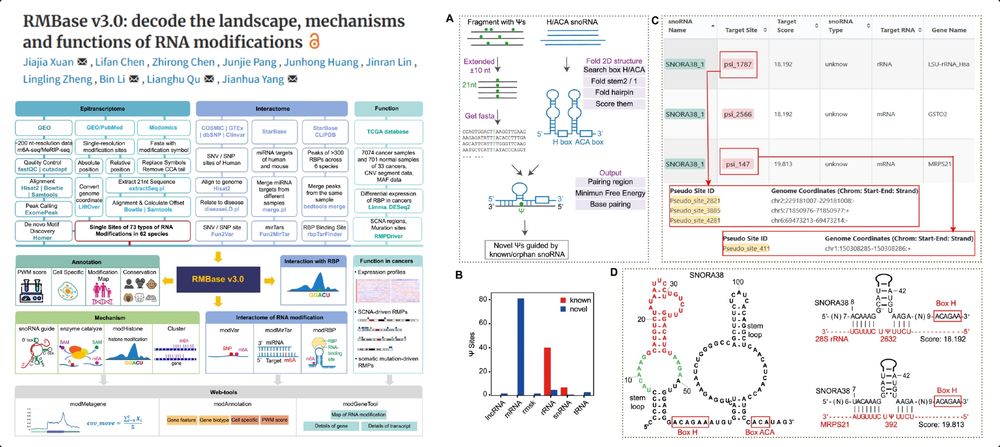
RMBase v3.0: decode the landscape, mechanisms & functions of RNA modifications
The Encyclopedia of RNA Epitranscriptome
More than m6A
"73 RNA modifications of 62 species"
bioinformaticsscience.cn/rmbase/
@narjournal.bsky.social 2024
academic.oup.com/nar/article/...
20.07.2025 19:06 — 👍 31 🔁 9 💬 1 📌 0
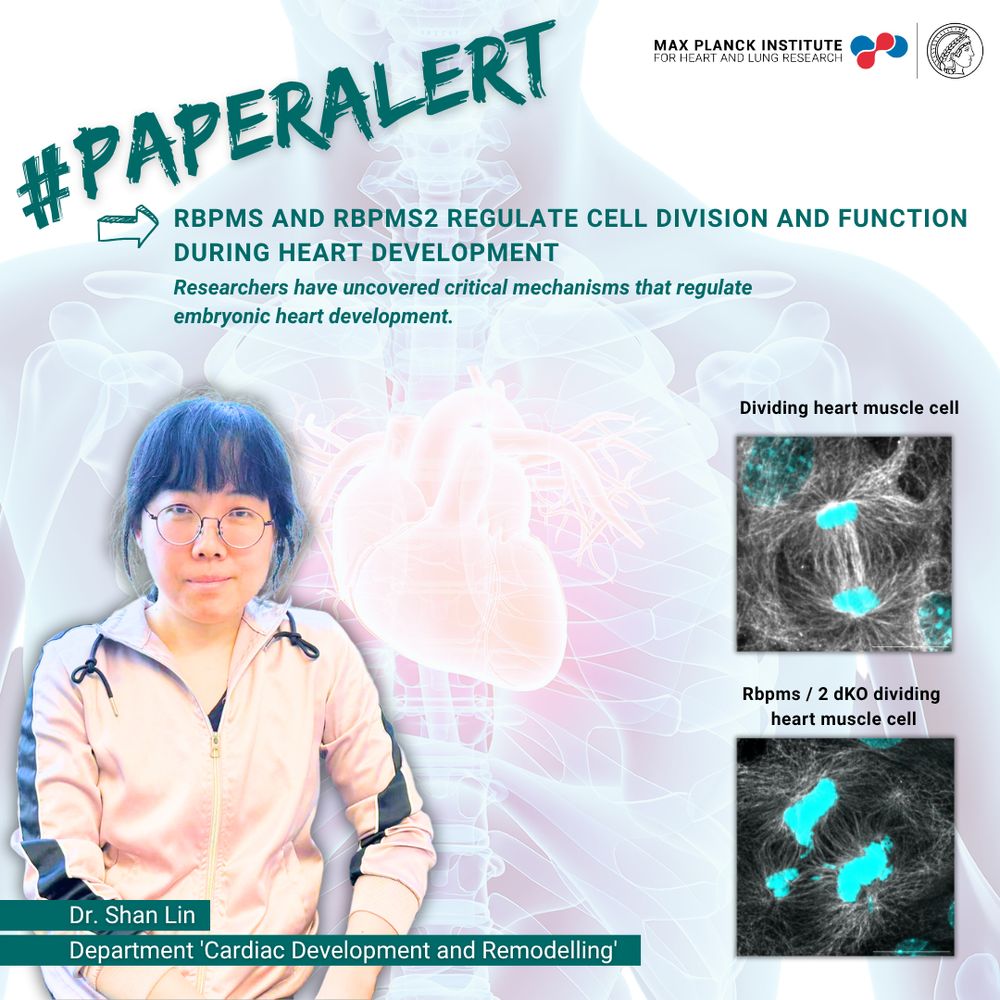
Scientists from our department 'Cardiac Development and Remodelling' discovered, that RBPMS and RBPMS2 regulate cell division and function during heart development, ensuring that the heart grows and matures correctly. Read the full paper in 'Developmental Cell' ➡️ lnkd.in/efwafKkE
17.07.2025 07:06 — 👍 1 🔁 1 💬 0 📌 0
Please consider eliminating your paywall on articles containing important information relevant to our current scientific crisis.
17.04.2025 20:39 — 👍 4 🔁 1 💬 0 📌 0

NSF halts grant awards while staff do second review
Action comes after DOGE team arrives at science agency
The National Science Foundation has put a cork in its grantmaking pipeline after billionaire Elon Musk’s Department of Government Efficiency set up shop at the agency this week. scim.ag/4iowX4t
17.04.2025 16:04 — 👍 51 🔁 31 💬 3 📌 1
Light sheet microscopy
Light sheet microscopy is a technology that enables near-isotropic imaging of large biological specimens over time. It opens new avenues to study biological processes with unprecedented imaging speed…
The @embo.org practical course on #lightsheet #microscopy is back!
Join us in #Dresden for 2 weeks with #your #samples, try the complete line-up of #lightsheet microscopes, tackle big data image analysis, sample prep, OA hardware & much more
Apply now!
Repost 🙏
meetings.embo.org/event/25-lsm
15.04.2025 08:32 — 👍 54 🔁 38 💬 1 📌 1
🤔
07.04.2025 16:22 — 👍 1 🔁 0 💬 0 📌 0
Wir sind ein gemeinnütziger Verein im hessischen Bad Nauheim, der im Bereich der Miniaturenstrategiespiele aktiv ist. Für uns sind dabei Gemeinschaft, Spielkultur und künstlerische Gestaltung wichtig.
EMBO/MSCA postdoctoral fellow at Francis Crick Institute in the Briscoe lab. Previously PhD Stainier Lab (heart dev) @MPI-HLR
Developmental biologist, lineage trajectories, cell fate decisions, cellular interactions, organ formation.
Disease Transcriptomics Lab, led by Nuno Barbosa-Morais, at the Gulbenkian Institute for Molecular Medicine (GIMM) in Lisbon, Portugal. @gimmfoundation.bsky.social
RNA biologist 🧬 | Postdoc @zarnack-group in JMU Würzburg | From wet lab roots to computational biology
🧬 Join our world-class courses, conferences, and workshops at the forefront of molecular life science and its applications. 👩🏻🔬 www.embl.org/events/ 🔬
Chair for Computational and Theoretical Biology
Computational Biology group located at Universtiy of Würzburg, Germany. We work on #RNA_splicing #RNA_modification #RNA_transport #RNA_stabiliy #RBPs_in_Disease #DNA_replication #DNA_double_strand_break
Neuro and Developmental biologist. PostDoc at day. Supervillain at night. He/him #BlackLivesMatter #TransRightsAreHumanRights
@mads100tist@mastodon.social
Your local ZEISS-Guy in Hesse.
Microscopy,... what else?
Our lab studies #development #regeneration #geneticrobustness @mpi_hlr. The account is run by various lab members.
Website- https://www.mpi-hlr.de/developmental-genetics
Danish developmental biologist based in Marseille - sometimes reading, sometimes sports, always 🇺🇦
Group Leader @CbiToulouse @Inserm | @ERC_Research | #zebrafish | #devbio & Regeneration | Cardiovascular disease
PhD student in @höferlab at MPI Marburg
The Höfer lab studies the epitranscriptomic mechanisms of gene regulation based on NAD-capped RNAs in bacteria and bacteriophages. We are located at the Max Planck Institute for terrestrial Microbiology, Marburg, Germany and area Member of SYNMIKRO
Passionate about uncovering how mRNPs form in the nucleus and how this intertwines with other aspects of gene expression. Based at Justus Liebig University Giessen, Germany.
https://www.uni-giessen.de/de/fbz/fb08/Inst/biochem/straesser/
Bioinformatiker, Skeptiker, Skeptix, Burgenfreak, Großfürst des Chaos
















