Beeswarm plot of the prediction error across different methods of double perturbations showing that all methods (scGPT, scFoundation, UCE, scBERT, Geneformer, GEARS, and CPA) perform worse than the additive baseline.

Line plot of the true positive rate against the false discovery proportion showing that none of the methods is better at finding non additive interactions than simply predicting no change.
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
04.08.2025 13:52 — 👍 125 🔁 56 💬 2 📌 6
#spatialproteomics #spatialbiology #multiplexedimaging #bioinformatics #python #scverse #opensource #singlecell #akoya #codex
05.05.2025 11:30 — 👍 1 🔁 0 💬 0 📌 0
𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬 offers:
✅ A unified API for segmentation, image processing, cell phenotyping, and spatial statistics
✅Consistent handling of shared dimensions across data structures
✅Built on xarray and dask for high flexibility and memory efficiency
✅Easy installation and usage
05.05.2025 11:30 — 👍 0 🔁 0 💬 1 📌 0
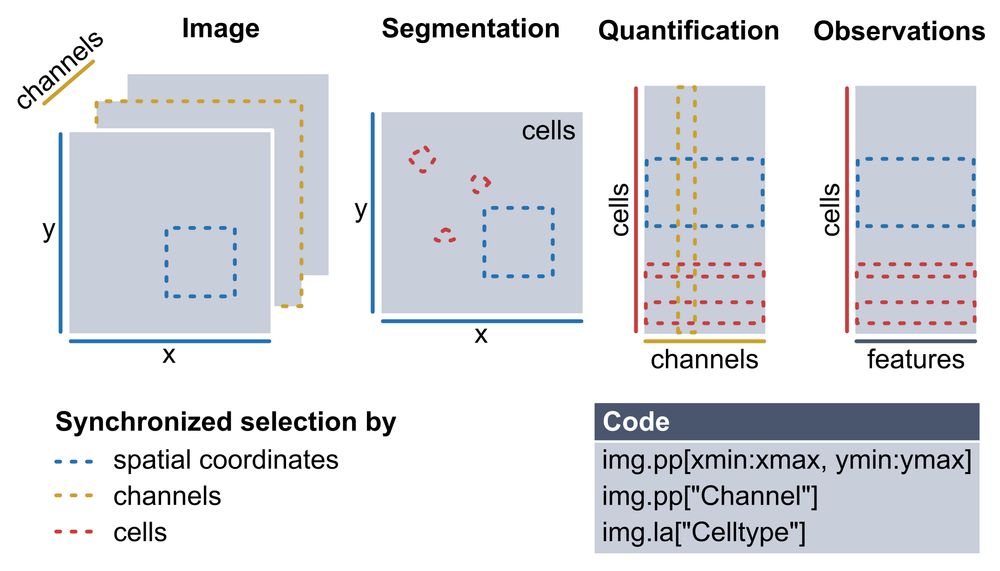
The spatialproteomics data structure enables synchronized subsetting across shared dimensions.
Multiplexed imaging (CODEX, MICS, IMC) gives single-cell resolution at the protein level — but analyzing these datasets requires stitching together many different tools and data structures.
You need to manage images, masks, expression matrices, and keep them all consistent.
05.05.2025 11:30 — 👍 1 🔁 0 💬 1 📌 0
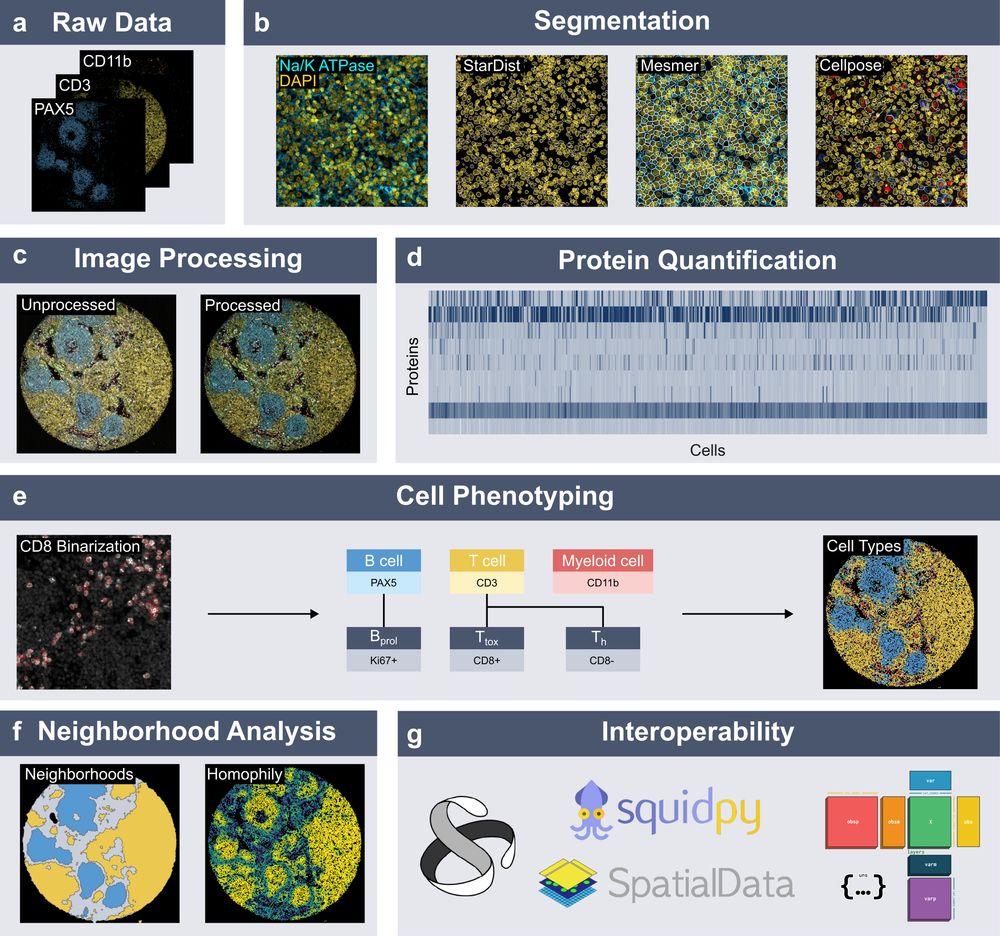
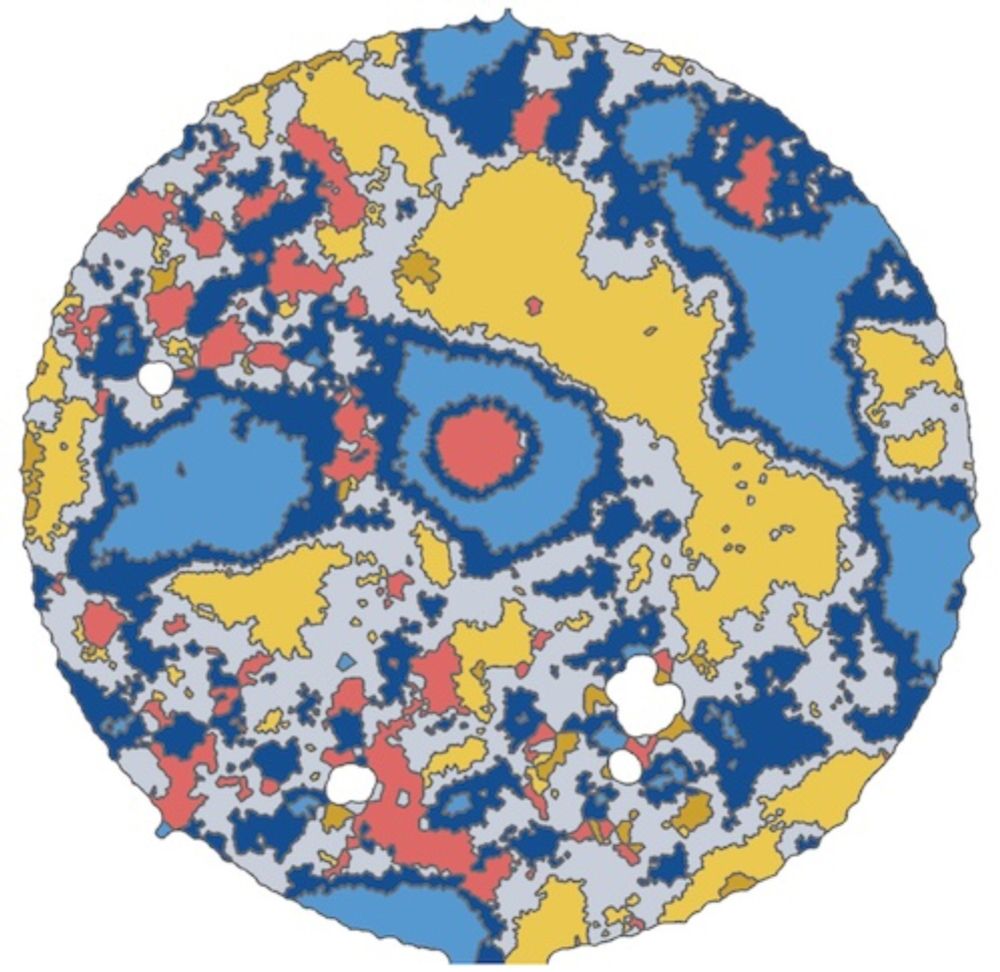
Spatialproteomics orchestrates workflows to analyze highly multiplexed images. It segments cells, processes images, quantifies proteins, predicts cell types, and provides neighborhood analysis methods, all while integrating into the scverse ecosystem.
New preprint out!
We introduce 𝐬𝐩𝐚𝐭𝐢𝐚𝐥𝐩𝐫𝐨𝐭𝐞𝐨𝐦𝐢𝐜𝐬, a Python package for end-to-end processing and analysis of highly multiplexed immunofluorescence imaging data.
Built on xarray and dask, with seamless integration into the scverse ecosystem.
www.biorxiv.org/content/10.1...
05.05.2025 11:30 — 👍 11 🔁 1 💬 1 📌 0
Spatialproteomics - an interoperable toolbox for analyzing highly multiplexed fluorescence image data https://www.biorxiv.org/content/10.1101/2025.04.29.651202v1
03.05.2025 23:47 — 👍 4 🔁 2 💬 0 📌 0
Mattermost is nice for communication, although it limits the file size, so maybe not ideal for sharing larger files. If you regularly share big files, maybe setting up ownCloud could be an option?
10.10.2023 20:31 — 👍 1 🔁 0 💬 1 📌 0
Medizinische Fakultät Heidelberg: Medical Data Scientist Programm
*Medical Data Scientist Postdoc* Program by Medical Faculty Uni Heidelberg. Join S.Dietrich, J.Lu & me to work on statistical& AI methods applied to single cell and spatial omics to improve immunotherapies:
www.medizinische-fakultaet-hd.uni-heidelberg.de/forschung/fo...
www.embl.org/about/info/m...
03.10.2023 12:59 — 👍 4 🔁 7 💬 2 📌 0
*The necessity and power of random, under-sampled experiments in biology...
*🍫-🐭, PERTURB-CAST, RUBIX...
👨🔬 Predoctoral Fellow at @EMBL.org as a member of the @borklab.bsky.social.
💻🧬 Using data science to analyze big bio-data.
🇪🇺 Part of the @embltrec.bsky.social expedition.
PhD candidate in BorkLab #embl #trec expedition team interested in #MobileGeneticElements #mobileDNA 🧬 & planetary #microbiome 🌎 mountaineer and climber 🧗♀️ ⛰️
PhD student @EMBL in the Krebs Lab
Computational medicine @ AU
Founder @ KH Biotechnology & medprofs
Interested in spatial omics, immunology, cancer & ageing
Find our software at: https://github.com/complextissue/
Personal: https://maltekuehl.com
Opinions my own.
PhD student in Statistical Bioinformatics at University of Zurich and SIB | Currently visiting EMBL
PhD student at EMBL-EBI & University of Cambridge
Postdoc Fellow at Saez-Rodriguez and Savitski labs (EMBL). Original from Córdoba, Spain.
We are an enthusiastic bunch of ELLIS fellows & scholars tackling the latest challenges in the application of #AI and #ML in the #LifeSciences @dkfz.bsky.social, @uniheidelberg.bsky.social, and @embl.org in Heidelberg!
Accessible AI Research | Open Source
Computational Biology | Pharmacology
Research Software Engineering
PI @ Helmholtz Munich (Computational Health)
@slolab.ai
Computational Cancer Genomics Group
EMBL's European Bioinformatics Institute (EMBL-EBI) provides open biological data resources and tools, and performs basic research in computational biology. https://www.ebi.ac.uk/
Head of Research and Development of the BioVisionCenter at the University of Zurich
An expedition to study coastal ecosystems and their response to the environment, from molecules to communities
PhD Student in the Zimmermann lab at EMBL Heidelberg 🦠
https://www.embl.org/groups/zimmermann/
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Guest Scientist IMP Vienna, Board of Directors NumFOCUS
Incoming Prof UMass Chan Medical
Previously Stanford Genetics, UW CSE.
physicist, bioinformatics / biostats researcher at University of Heidelberg
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing