PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
The final paper of my UoS postdoc is out now in PNAS! How to detect developmental plasticity in the fossil record:
Featuring
@tomezard.bsky.social
@jamesmulqueeney.bsky.social
@katsamenis.bsky.social
@clivetrue.bsky.social
@thefosterlab.bsky.social
among others
www.pnas.org/doi/10.1073/...
07.07.2025 10:41 — 👍 20 🔁 6 💬 1 📌 0
Some great work by @aniekebrombacher.bsky.social! Super cool to be part of this collaborative project!
03.07.2025 23:09 — 👍 1 🔁 0 💬 0 📌 0
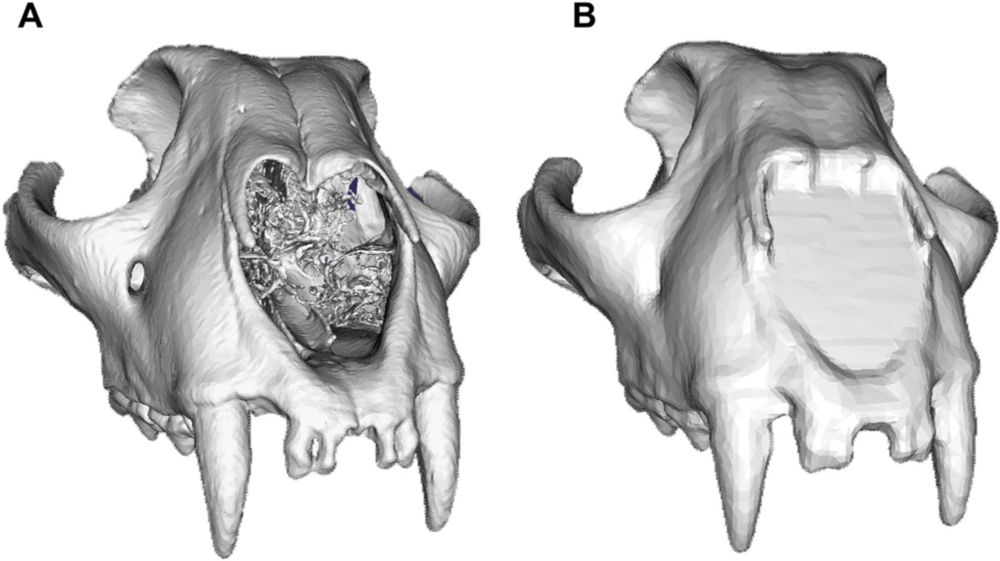
New paper led by @aniekebrombacher.bsky.social using x-ray CT and laser ablation to detect plastic environmental responses in fossil individuals www.pnas.org/doi/10.1073/pnas.2421549122 featuring @jamesmulqueeney.bsky.social @clivetrue.bsky.social @thefosterlab.bsky.social
03.07.2025 21:53 — 👍 22 🔁 15 💬 1 📌 1
No worries. I have been working with these methods for a while so good to try and demonstrate their potential and help others utilise them. Best of luck with things!
27.04.2025 14:14 — 👍 2 🔁 0 💬 0 📌 0
Overall, we show the landmark-free methods like Deterministic Atlas Analysis can yield correlated results with traditional manual landmarking & semilandmarking methods. We use the results to propose how to improve these methods in the future!
27.04.2025 12:16 — 👍 5 🔁 0 💬 0 📌 0
Finally, we compare the downstream estimates of morphological disparity, evolutionary rates and phylogenetic signal. Here, we find significant correlations between methods for almost all metrics.
27.04.2025 12:15 — 👍 4 🔁 0 💬 1 📌 0
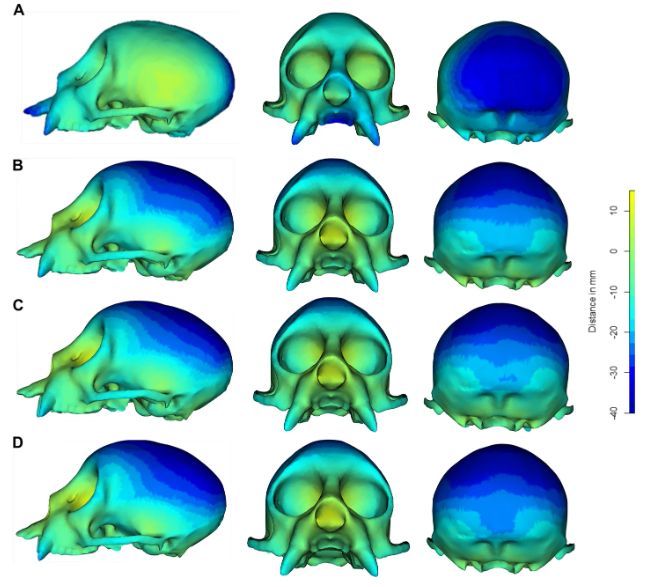
We then compare heatmaps of how shape is measured using each method. To do this we compare the shape of different crania with estimated mean shapes of each analysis (landmark top, DAA below). Here is shown for the specimen, Cacajao calvus.
27.04.2025 12:14 — 👍 2 🔁 0 💬 1 📌 0
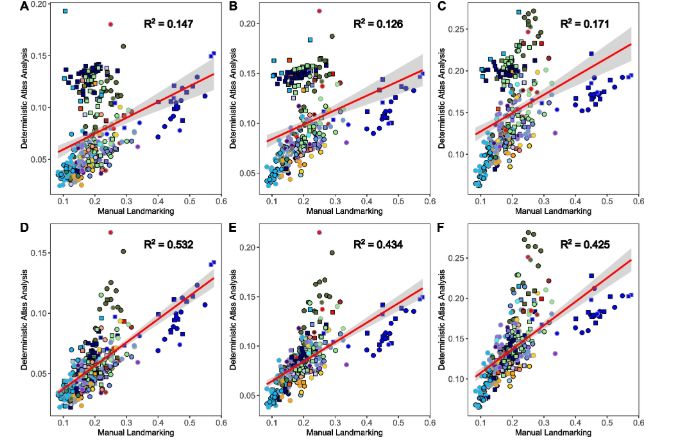
We then compare the correlation between the two methods using Euclidean distances, the Mantel test and the Procrustean randomisation test. We find the methods correlate and that using Poisson meshes significantly improves the correlation between methods.
27.04.2025 12:13 — 👍 2 🔁 0 💬 1 📌 0
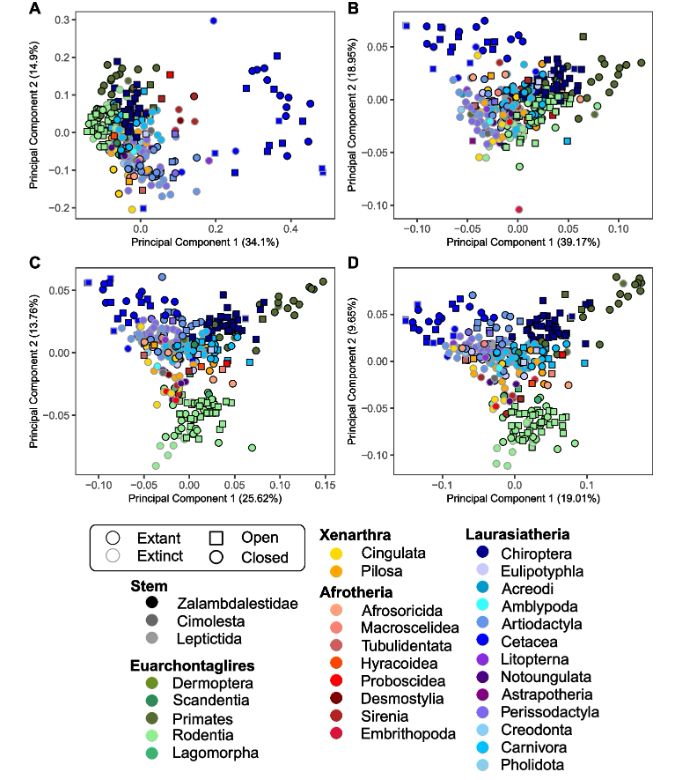
We then visually compare the results of the manual landmarking analysis (754 landmarks and semilandmarks; top left) with the landmark-free approach using 45, 270 and 1,782 control points (top right to bottom left). We notice differences mainly for Cetacea and Primates.
27.04.2025 12:12 — 👍 2 🔁 0 💬 1 📌 0
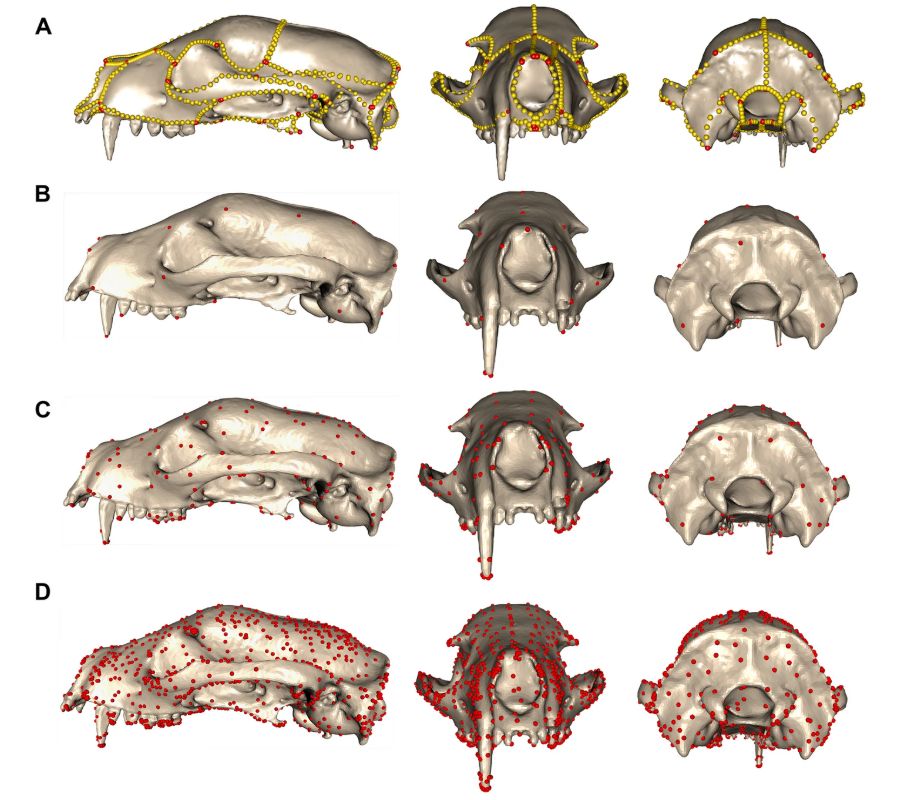
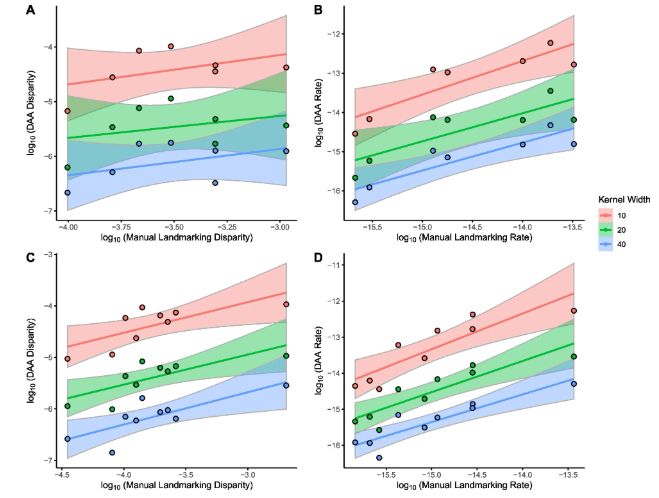
Once overcoming modality issues, we apply the method to 322 placental mammal crania as seen in Goswami et al. 2022 (science.org/doi/full/10....). Here, we apply and test a range of kernel widths, which changes the number of control points used to compare shape (replacing landmarks).
27.04.2025 12:11 — 👍 2 🔁 0 💬 1 📌 0
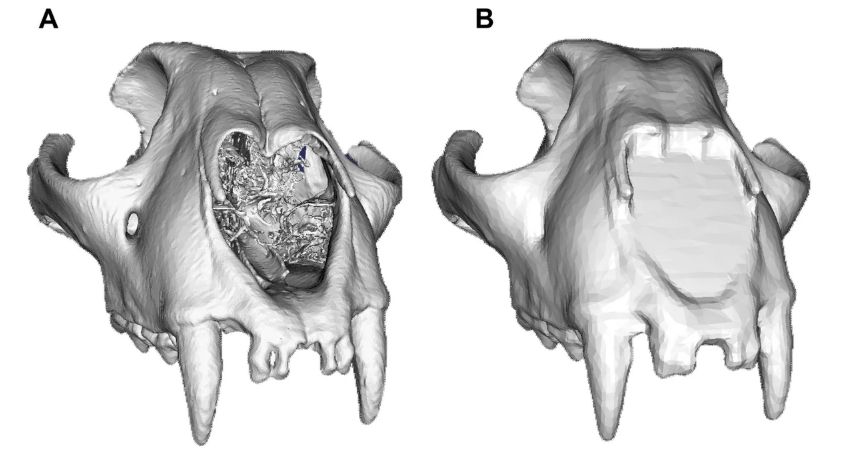
We focus on comparing an implementation of Large Deformation Diffeomorphic Metric Mapping (LDDMM) called a Deterministic Atlas Analysis (DAA) with manual landmarking techniques. We first notice that data modality affects the results, which we overcome by making Poisson meshes.
27.04.2025 12:08 — 👍 3 🔁 0 💬 1 📌 0
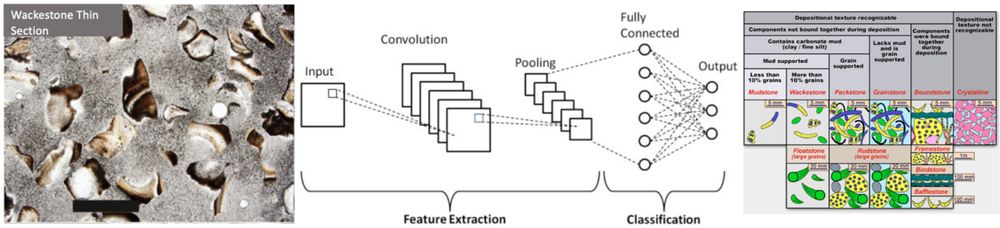
In our March Newsletter we hear from UK-IODP scientists Cedric John, Lewis Grant & @jamesmulqueeney.bsky.social, who've each developed novel applications of machine learning to facilitate rapid, accurate classification of IODP samples
issuu.com/uk-iodp/docs...
@ecord.bsky.social @anzic.bsky.social
25.03.2025 15:03 — 👍 5 🔁 2 💬 0 📌 0

a 3d rendering of the skull and cranial muscles of the Eurasian oystercatcher (Haematopus ostralegus)
turns out you can do some pretty cool stuff with the power of diceCT, SPROUT (www.biorxiv.org/content/10.1...), and the SmARTR pipeline (www.sciencedirect.com/science/arti...)
21.12.2024 15:28 — 👍 40 🔁 10 💬 0 📌 0

Reminder of the January 8th deadline for this reef palaeoecology PhD project at @sotonoceanearth.bsky.social with @tomezard.bsky.social + @chrisgoatley.bsky.social :)
www.findaphd.com/phds/project...
18.12.2024 14:21 — 👍 14 🔁 14 💬 1 📌 0
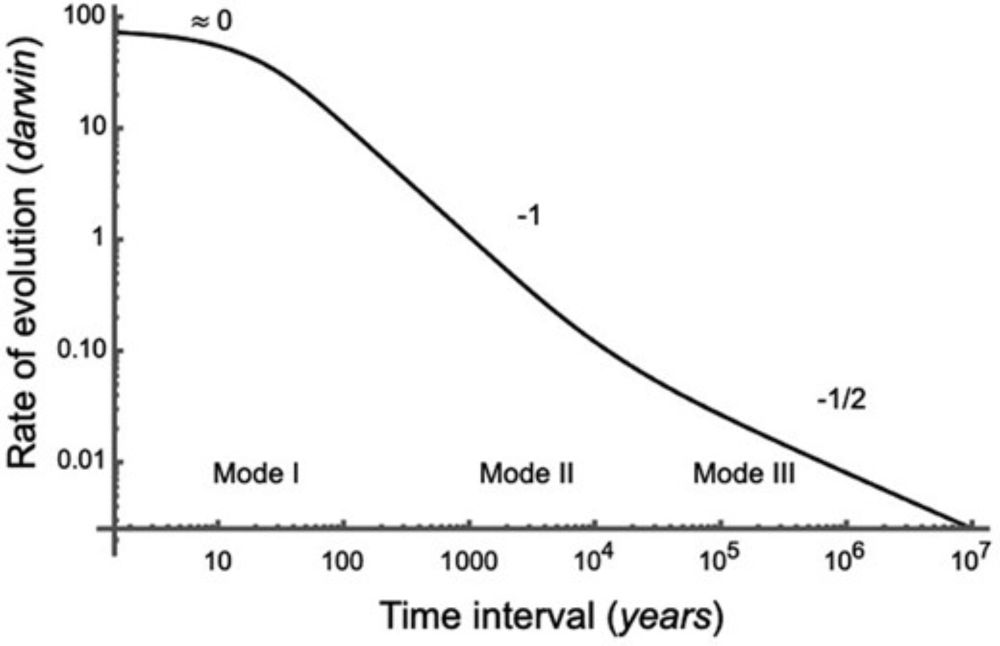
Three modes of evolution? Remarks on rates of evolution and time scaling
Abstract. Rates of evolution get smaller when they are measured over longer time intervals. As first shown by Gingerich, rates of morphological change meas
Highly recommend this very interesting new paper. I'll be reading it again--and then probably again & again. It's insightful, idea rich, & thought provoking even in places where I disagree w/ interpretations! 🧪 #EvoSky
"Three modes of evolution? Remarks on rates of evolution and time scaling"
11.12.2024 16:58 — 👍 38 🔁 18 💬 0 📌 0
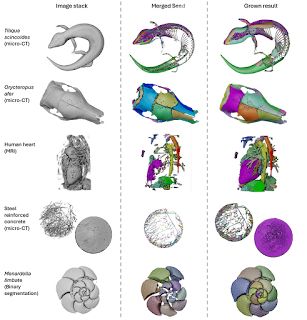
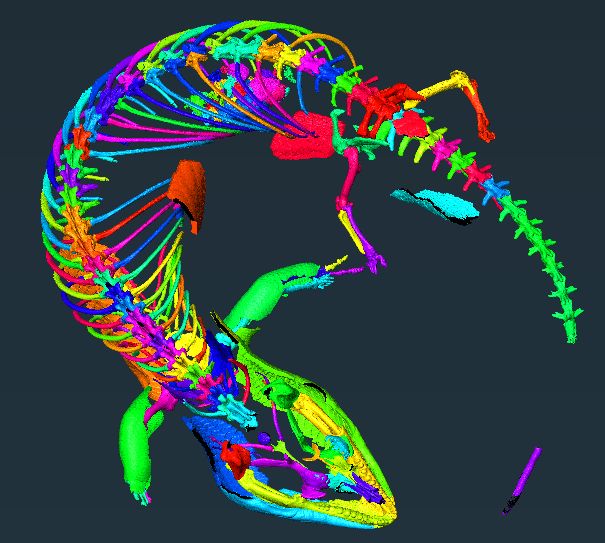
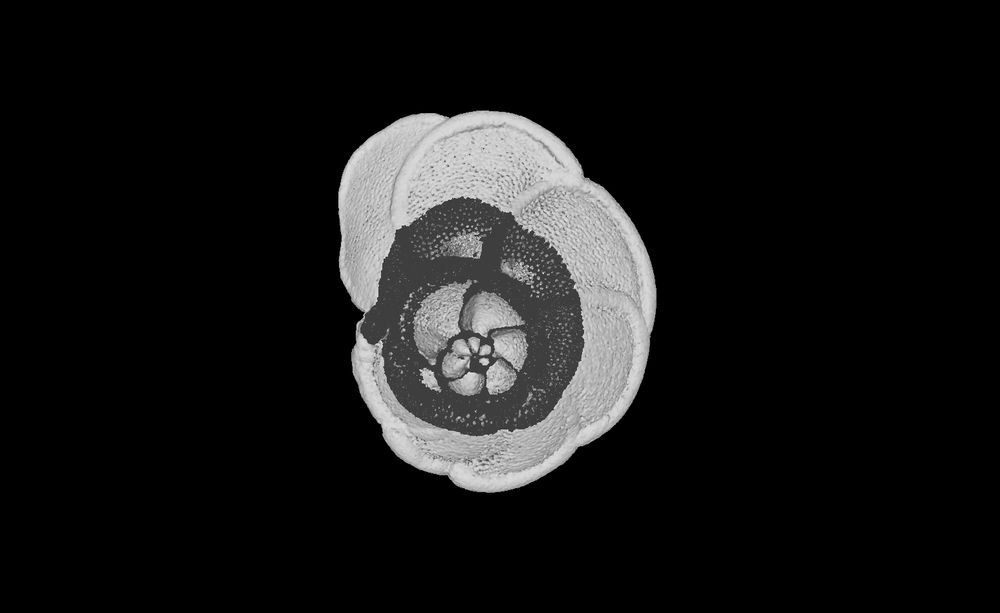
Figure showing diverse datasets analysed with SPROUT, including a skink skeleton, aardvark skull, human heart, concrete block, and foraminifera.
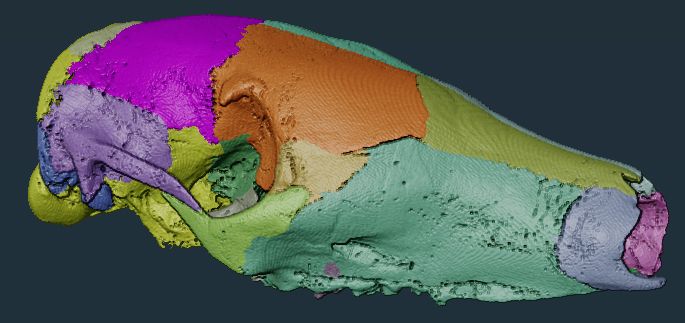
New paper! For all of you working with 3d scans (e.g. micro-CT, MRI), check out SPROUT, a rapid open-source tool for generating segmented and parcellated data, meaning your scans are separated into the individual elements without any manual labelling or training. www.biorxiv.org/content/10.1...
24.11.2024 13:09 — 👍 213 🔁 92 💬 10 📌 6
Our new paper on #AI for evolutionary morphology is out! This massive team effort covers the history of AI for studying morphology, reviews new tools, provides many case studies & a prospectus for using AI to progress diverse topics in evolutionary morphology. academic.oup.com/iob/article/...
09.11.2024 13:41 — 👍 10 🔁 7 💬 0 📌 0
I am a paleontology enthusiast who loves ceratopsians, stegosaurs, and Mesozoic birds.
Pfp by Fabrizio Lavezzi
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
Functional morphologist, particularly in #Carnivora and #Reptilia.
#ecomorphology #macroevolution
Currently on the postdoc hunt!
Institute of Evolutionary Biology @BiolUW @UniWarszawski. From molecules to bones, we are committed to the study of evolution, phylogenetics, and taxonomy.
Research Fellow at @biology.ox.ac.uk interested in the evolution of plants and algae
✒Assistant Professor of Geology, Jogamaya Devi College, University of Calcutta. 🌏PAGES ECN Steering Committee Member.
🐚Loves foraminifera, pteropods and photography. 🔬Ph.D. from IIT Kharagpur.
Assistant Professor @Princeton EEB. #phylogenetics/#macroevolution/#paleobiology
http://simoes-lab.com
Posts and views are of my own.
SCIMPI is an inexpensive subsea borehole observatory for unconsolidated sediments. Our latest deployment is IODP³-NSF/ECORD Expedition 501: New England Shelf Hydrogeology.
Tell us which sensors YOU would like on a SCIMPI.
www.TranscendEngineering.com
Diatoms, algae, paleolimnology, limnology, freshwater ecology 💧🔬| Field stations, science education, educational board games | Associate Professor and E&O Director of field station in Texas | Travel 🗺, cross stitch 🪡, highland dance and clogging 🩰
she/her
A bit of modeling, a bit of experiments, lots of evolution. Researcher
@cnrs @cefemontpellier
Lecturer in Biomedical Imaging | Biomedical Imaging and X-ray Histology lead at the μ-VIS X-ray Imaging Centre (muvis.org)
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
Theoretical & Quantitative Ecologist. Dad. Whisky lover. Football coach. Relearning the drums. Not necessarily in that order.
See also @theorecol@ecoevo.social
He/him
Scientist Interested in all things oceans and climate (esp. carbon cycle). Bike rider, hiker and nature lover. Privileged white feminist. Advocate for diversity.
Mexican Historian & Philosopher of Biology • Postdoctoral Fellow at @theramseylab.bsky.social (@clpskuleuven.bsky.social) • Book Reviews Editor for @jgps.bsky.social • https://www.alejandrofabregastejeda.com • #PhilSci #HistSTM #philsky • Escribo y edito
Peter Buck Postdoctoral Fellow at the Smithsonian Museum of Natural History
Paleontologist and proud Cajun. Love fossils, phylogenetics, and reptiles. (she/her).
Cooking adventures on instagram @cook_with_kels
Postdoctoral scholar, University of Chicago, Tsegai lab | I study mammalian locomotor evolution | I approach my research using integration, biomechanics, and phylogenetic methods.
LaBella Lab studies codon usage (aka "silent" genetic changes) and evolution from humans to fungi | Bioinformatics & Genomics | UNC Charlotte
Run by Abbe LaBella
Opinions are my own and do not reflect any affiliated organization.
Associate Professor @ University of Kentucky. Evolution / Ecology / Math. Biker. Aikidoka. #BlackLivesMatter
Editor in Chief of Theoretical Population Biology
http://vancleve.theoretical.bio