mailejim.bsky.social
This work is a four-year journey that would not be possible without my amazing partners @mailejim.bsky.social, Mounica Vallurupalli, and Kai Cao, and other co-authors. And thank you to my PI Fei Chen, the Golub Lab, @broadinstitute.org, @harvardmed.bsky.social, for your support! (10/10)
06.11.2025 18:59 — 👍 0 🔁 0 💬 0 📌 0
Our work shows that RNA splicing can be harnessed as a new modality for cell type-specific gene regulation, unlocking new possibilities for gene therapy and precision medicine. It also highlights how high-quality experimental data powers AI models. (9/10)
06.11.2025 18:59 — 👍 1 🔁 0 💬 1 📌 0
We further applied SPICE to design sequences that specifically target cancer cells carrying splicing factor mutations. We identified synthetic sequences that are selectively spliced only in cells with RBM5 or RBM10 mutations, which are commonly seen in lung cancers. (8/10)
06.11.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0

To go beyond prediction, we built Melange, a generative model that designs new sequences with programmed cell type-specific splicing. We experimentally validated that Melange can design sequences that splice only in neural-lineage cells, and not cells from other lineages! (7/10)
06.11.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0
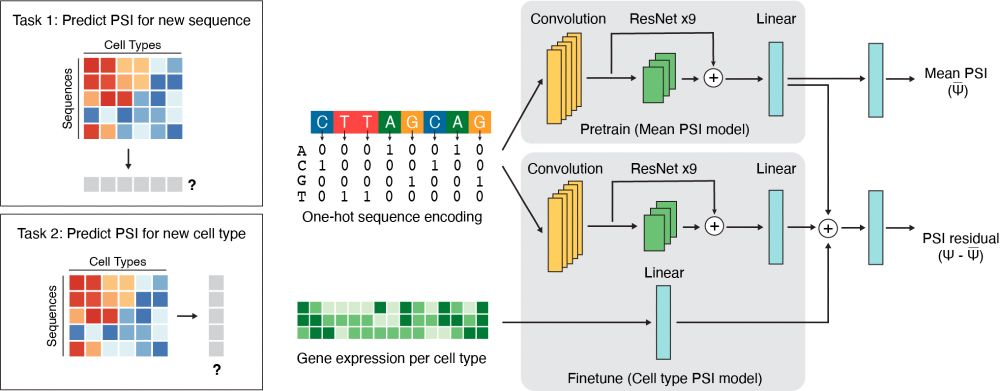
Using this dataset, we built Soma, a deep learning model that predicts how any RNA sequence will splice across cell types.
We drew inspiration from ChromBPNet (@anshulkundaje.bsky.social) to encode cell type-specific information directly from gene expression data. (6/10)
06.11.2025 18:59 — 👍 2 🔁 0 💬 1 📌 0
We profiled 46,000+ sequences across 43 cell lines spanning 10 lineages – the most diverse set of cell types ever tested in an MPRA (>10x more than previous studies) – and uncovered widespread cell type–specific splicing events. (5/10)
06.11.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0

Good ML models require high-quality experimental data, but large-scale, cell type-resolved data on RNA splicing is limited. Hence, we built SPICE (Splicing Proportions In Cell types), an integrated experimental + AI framework for the generative design of cell type-specific RNA sequences. (4/10)
06.11.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0
Beyond existing cell type targeting strategies like promoters, enhancers, or viral vectors, we asked: can we design RNA sequences that use alternative splicing as an underexplored biological process to control cell type-specific gene expression? (3/10)
06.11.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0
Every cell in your body contains the same DNA, but different cell types – like neurons or cancer cells – perform vastly different functions. Being able to turn genes on or off in specific cell types is key for understanding biology and for building precise, safe gene therapies. (2/10)
06.11.2025 18:59 — 👍 1 🔁 0 💬 1 📌 0
Thank you to the award committee, my PI Fei Chen and my lab, all my mentors, collaborators, family and friends for your support!
04.03.2025 18:37 — 👍 1 🔁 0 💬 0 📌 0
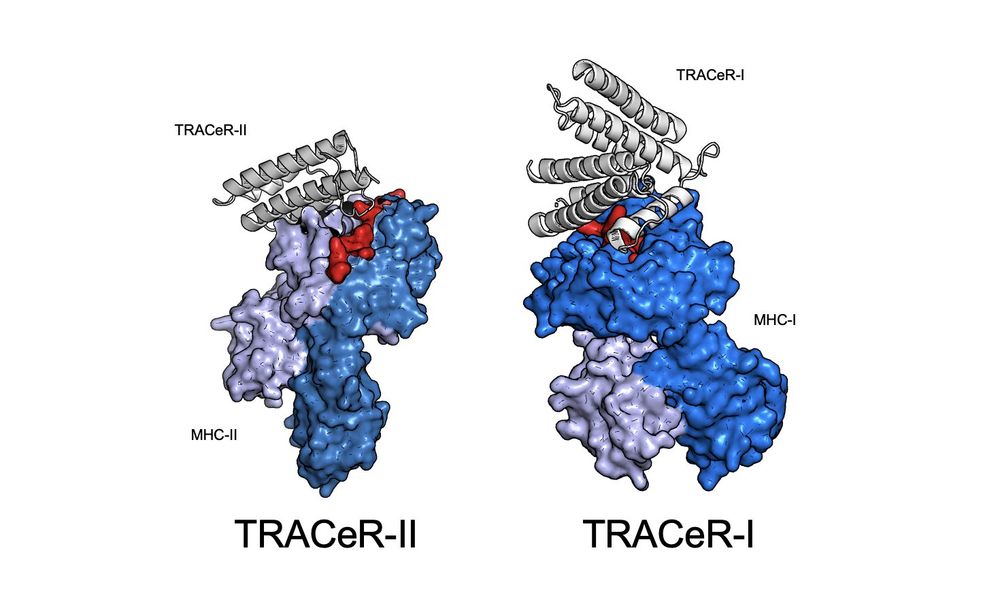
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
17.12.2024 00:56 — 👍 92 🔁 29 💬 2 📌 9
So at the beginning of my postdoc I struggled to amplify Chlamy genes (easily 75%+ GC) and did pretty much the same comparison and found the same results
DMSO= helps but kills pol processivity
Betaine= Lots of spurious products
1,2-propandiol= processivity retained and usually specific product
12.12.2024 07:00 — 👍 24 🔁 4 💬 0 📌 0
Dr. Margaret Oakley Dayhoff
I took biochem in 2001, and for nearly 20 years read amino acid sequences daily… and I never knew Dayhoff named them or even the logic behind things like Q until last Friday (h/t Mike Janech). Also, this is another big Dayhoff moment for me. She was incredible!
#proteomics #bioinformatics
24.11.2024 12:39 — 👍 198 🔁 79 💬 14 📌 7
Nah you're still a cow! Mooooo.
21.11.2024 17:47 — 👍 0 🔁 0 💬 0 📌 0
@mjafreeman.bsky.social's BlueSky starter packs for 'mechanistic' biologists
Here’s my spreadsheet of starter packs (>80!) related to broadly ‘mechanistic biology’ plus some intriguing extras
Complete with collective nouns
I’ve been tracking these but now can’t keep up
Hope it’s helpful
2/2
docs.google.com/spreadsheets...
19.11.2024 21:45 — 👍 204 🔁 123 💬 39 📌 16
Iranian-Scottish Londoner lost in the East, trying to get his head around scientific data and publishing
Assistant Professor of Biochemistry at Stanford. Previously at MIT, Broad Institute, and Harvard Society of Fellows. Microbiology and biotech 🧬 https://gaolab.bio/
Working for a diverse, global and multidisciplinary scientific community focused on the cell, the basic unit of all life.
Making Foundational Discoveries @fredhutch.org
fredhutch.org/basicsciences
Chemist at HHMI's Janelia Research Campus. Passionate about designing, building, and giving away fluorescent dyes to illuminate biological systems. Striving to be positive about all things chemistry (except ChemDraw).
ORCID: 0000-0002-0789-6343
Our lab uses experimental and computational methods to design de novo proteins | @Stanford
Prof. Breaker is an RNA biochemist, HHMI Investigator, Sterling Professor, and Chair of Molecular Biophysics and Biochemistry at Yale University, New Haven, CT.
Professor of Neurogenetics, Centre for Neural Circuits and Behaviour, University of Oxford. Genetic dissection of Sexual Behaviour.☘️ Drosophila neuroscience, genetics, genomics, sexual behaviour
http://www.dpag.ox.ac.uk/research/goodwin-group-1
🌍Direct DNA/RNA analysis for anyone, anywhere
🧬Short to ultra-long reads in real-time for rapid insights
🔬Products for research use only
Editor in Chief of Nature, geneticist, editor, accidental potter. All views my own
Synthetic DNA for Health & Sustainability
Global Alliance for Spatial Technologies: an organized whole that is perceived as more than the sum of its parts. Democratizing Spatial-Omics Techs
Scientist. Single Cell Genomics Group Leader at CNAG. Co-founder & CSO at Omniscope. ICREA Professor.
#Scientist🧑🔬 at #Illumina
✶ All things Bio-Omics-Informatics aka Bio-Omician
✶ Alum of #UTSA #sfu #HydUniv #CSIRCIMAP
✶ Celebrate 🏏 #Cricket & ♟️ #chess
✶ Exploring globe 🌍 through #workation 🇮🇳>🇨🇦>🇪🇸>🇺🇸
#linkedin priyankamishra17
features editor @endpts.com / fall 2021 fellow
@ksjatmit.bsky.social
Writing (and editing) stories on rare disease medicine, diagnostics and China
Reach out privately on Signal: jaredwhitlock.73
Assoc. Prof. Aalborg University 🇩🇰 Applied microbial ecology with focus on wastewater treatment, anaerobic digestion, and soil. HQ MAGs, transcriptomics, rRNA operon sequencing, culturomics, biofilms, EPS, and functional amyloids.
Biochemist 🧪| Empowering Life Scientists and Science Communicators to Achieve Their Career Goals | UCLA PhD + NIH Alum | Pharma R&D @GSK + @AstraZeneca 💊 | #SciComm Director ✍️| Career Coach | Star Trek | Sherlock Holmes | 🎹 | https://larrymillerphd.com
Researcher at Google DeepMind in London. Previously PhD at Cambridge University.
Professor, Santa Fe Institute. Research on AI, cognitive science, and complex systems.
Website: https://melaniemitchell.me
Substack: https://aiguide.substack.com/
Professional Stumbler. Shenanigans with Earth Science & Machine Learning
Visiting researcher at google & assistant prof at it:u (at)







