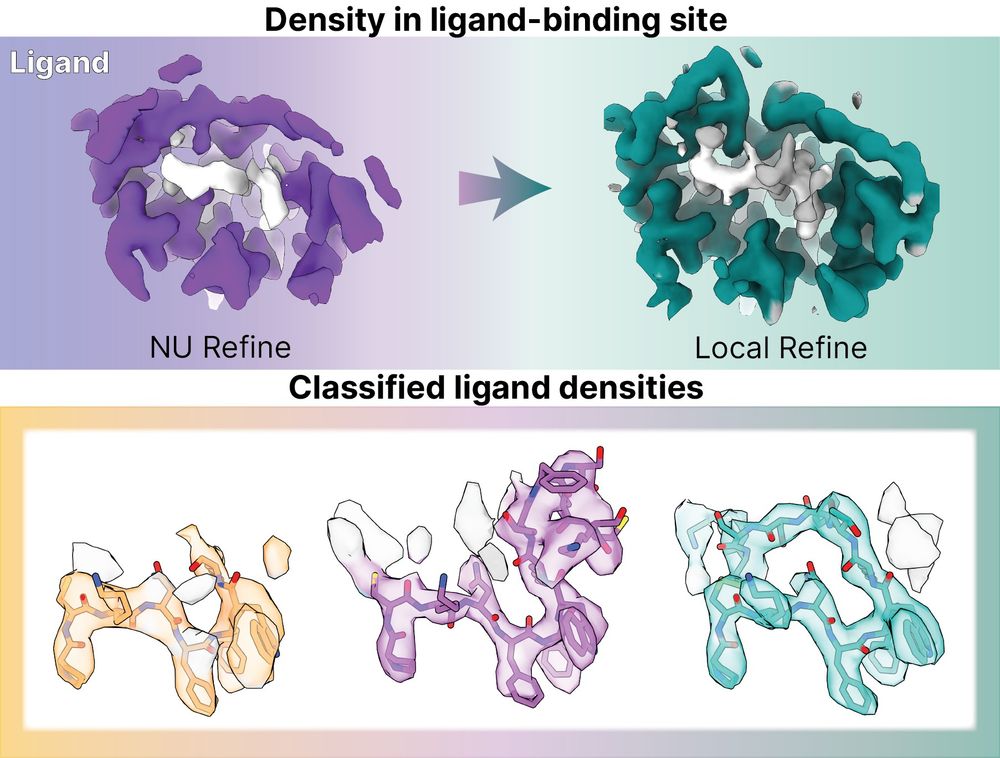
Grappling with poorly-resolved ligand binding sites in #cryoEM maps?
Check out our new case study on a peptide ligand-bound GPCR showing how practical tips and features in #CryoSPARC v4.7 improve locally refined map quality and reveal clearer ligand density!
guide.cryosparc.com/processing-d...
29.04.2025 15:22 —
👍 32
🔁 7
💬 0
📌 0
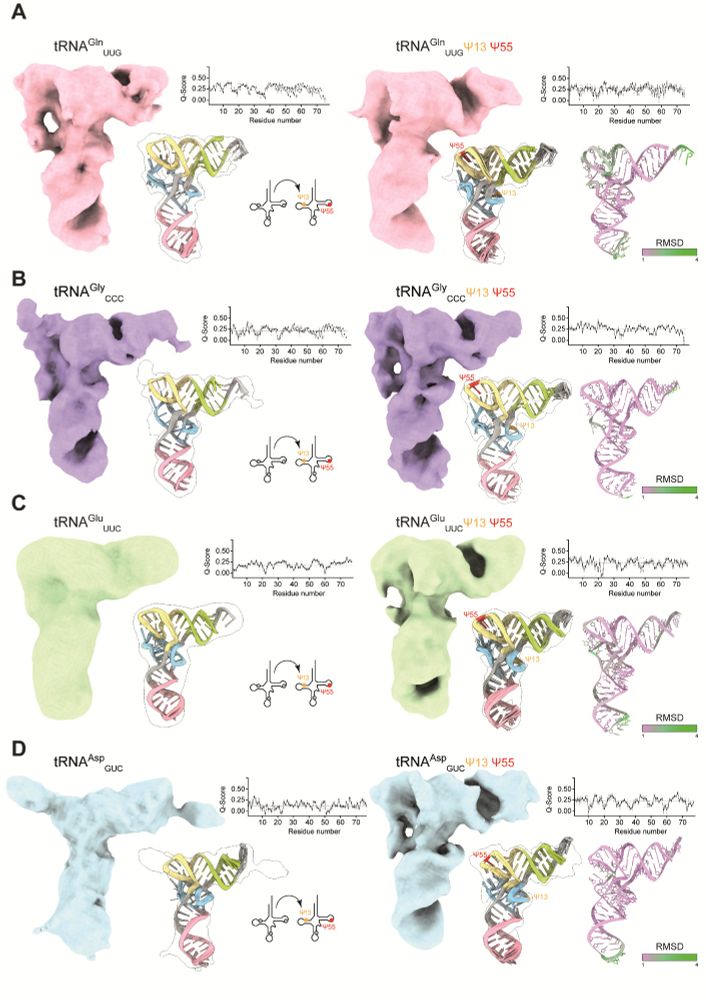
Our work is the first study to provide structures of several tRNAs before and after the introduction of modifications at specific sites. In detail, we analyzed 4 human tRNA species to decipher structural and stability effects induced by the installation of Ψ at single or multiple sites.
30.04.2025 04:11 —
👍 2
🔁 1
💬 1
📌 0
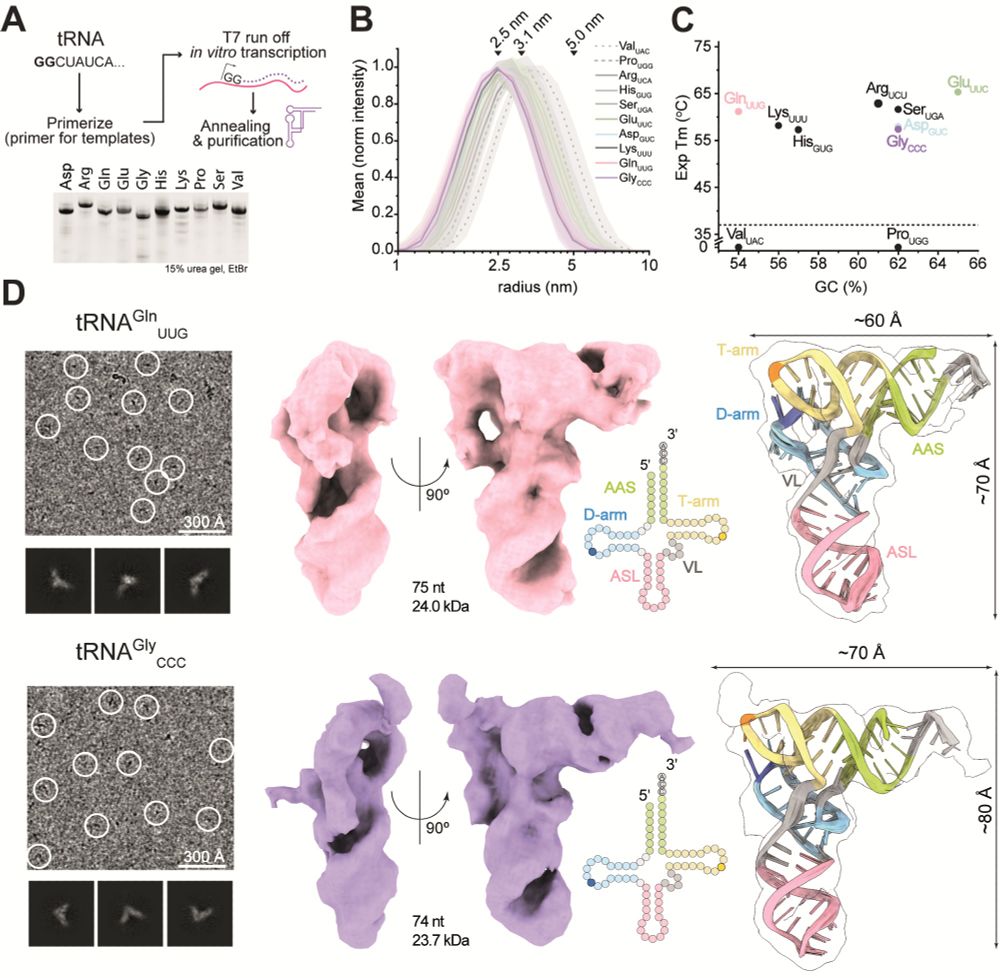
We used single particle cryo-EM to directly visualize several human tRNAs and analyze the effects of individual tRNA modifications. To our knowledge, the work presents interpretable maps of the smallest asymmetric particles analyzed by single-particle cryo-EM so far.
30.04.2025 04:11 —
👍 4
🔁 1
💬 1
📌 0
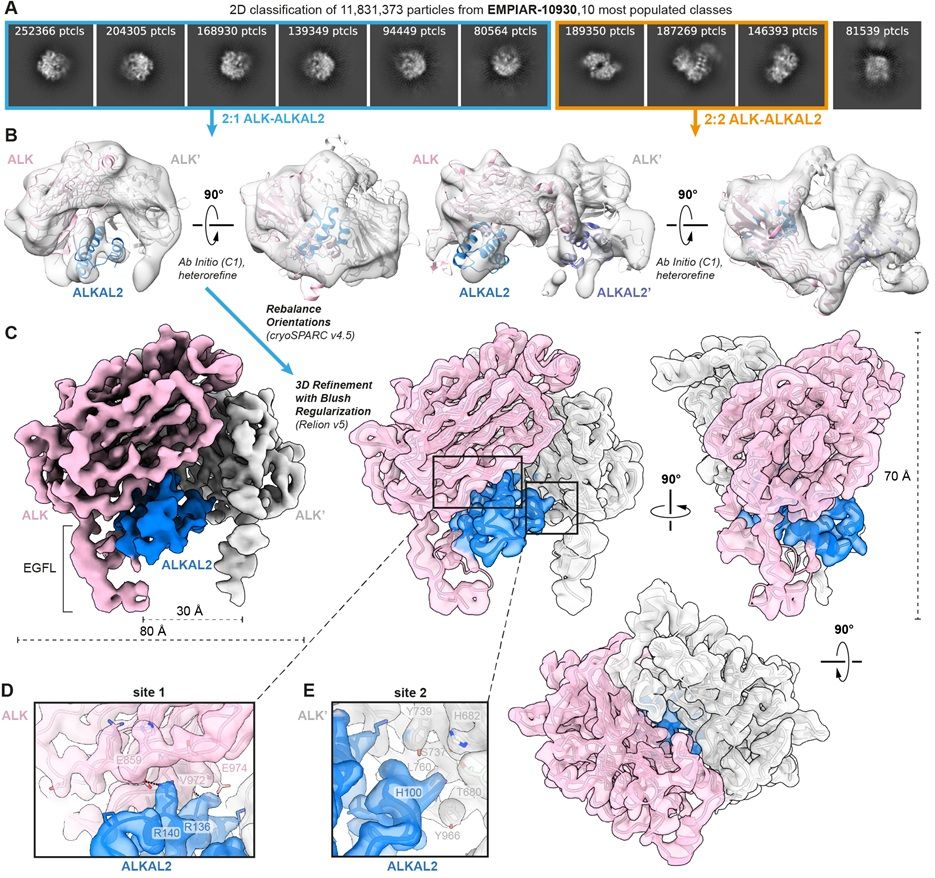
Reanalysis of cryo-EM data from EMPIAR entry EMPIAR-10,930 deposited by Reshetnyak and colleagues, Nature (2021). (A) Ten most populated 2D class averages after performing 2D classification on 11.8 million selected particles. Classes corresponding to 2:1 ALK–ALKAL2 particles are highlighted in blue, while classes corresponding to 2:2 ALK–ALKAL2 particles are highlighted in orange. (B) Ab initio model generation in cryoSPARC results in classes corresponding to both a 2:1 ALK–ALKAL2 and 2:2 ALK–ALKAL2 assembly. Hetero-refinement of the 2:1 and 2:2 ALK–ALKAL2 classes results in the two models displaying 2:1 and 2:2 ALK–ALKAL2 assemblies, respectively. The ALK–ALKAL2 complexes with 2:1 (PDB 7nwz) and 2:2 (PDB 7n00) stoichiometries are shown fitted inside the maps, with ALK colored pink/gray and ALKAL2 colored blue/purple. (C) 3D refinement; a front view is shown of a map sharpened in RELION using a B-factor of −100 Å2, with ALK colored pink/gray and ALKAL2 colored blue. Front, side, and top views are shown of a transparent map, sharpened using deepEMhancer, with fitted structural model.
Reports of diverse stoichiometries have complicated our understanding of the complex between ALK kinase and its #cytokine ligand ALKAL2. @janfelix.bsky.social @savvideslab.bsky.social &co reanalyze #cryoEM data to clarify the situation @plosbiology.org 🧪 plos.io/4lvRfvu
15.04.2025 13:55 —
👍 8
🔁 3
💬 0
📌 0
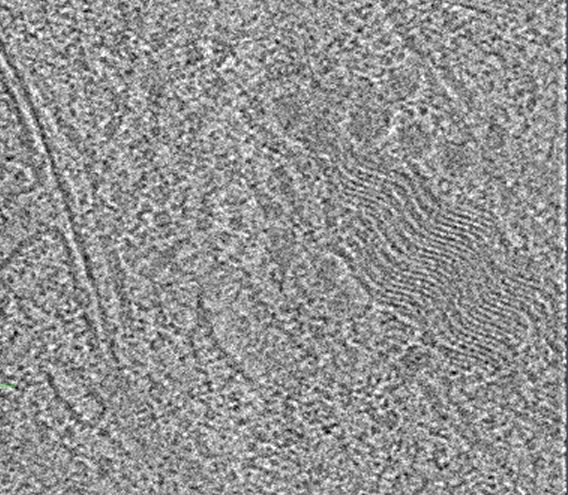
Does anyone have any idea what this wavey guy is? 🌊🌊🌊🌊#TeamTomo #CryoET
In-situ tomogram of milled RPE-1 cells.
07.03.2025 16:02 —
👍 18
🔁 4
💬 1
📌 0
Fast event-based electron counting for small-molecule structure determination by MicroED pubmed.ncbi.nlm.nih.gov/39982366/ #cryoEM
22.02.2025 21:58 —
👍 2
🔁 1
💬 0
📌 0
13.02.2025 04:37 —
👍 0
🔁 0
💬 0
📌 0
Interested in #CryoEM structure refinement?
Then this #webinar is for you: "Cryo-EM structure refinement with density-guided simulations in #GROMACS"
🗓️ 4 March 2025, 15:00 CET
Registration & further info ➡️ bit.ly/41b6L7W
11.02.2025 09:07 —
👍 10
🔁 3
💬 0
📌 1
Nice - first CryoEM structure of ALK2 in complex with a ligand! #BMP6 #ACVR1
11.02.2025 11:56 —
👍 8
🔁 1
💬 0
📌 0
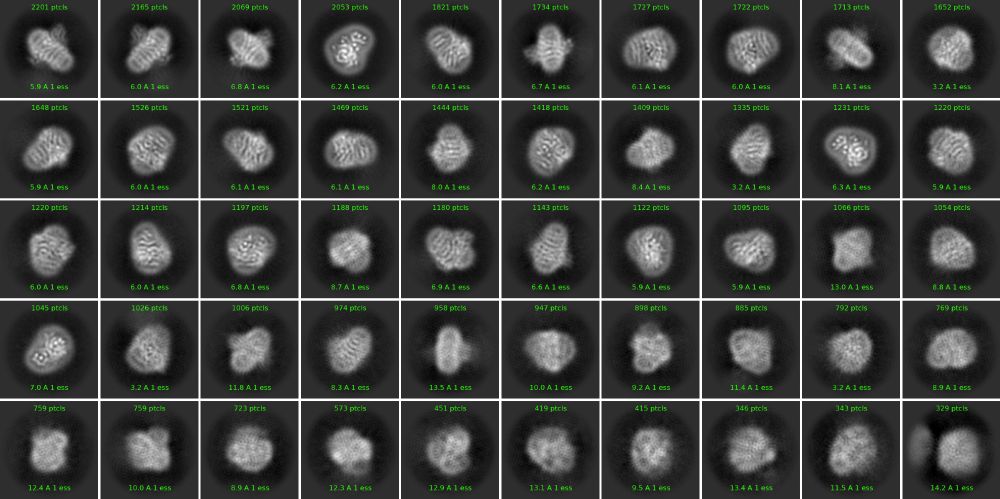
Mystery Protein Challenge Calling all structural biology enthusiasts and #cryoEM aficionados!
Can you identify the protein shrouded in this 2D class average?
Hint:
This protein plays a vital role in the respiratory chain.
11.02.2025 13:49 —
👍 3
🔁 1
💬 2
📌 0
Structure of the F-tractin–F-actin complex | Journal of Cell Biology | Rockefeller University Press
This study reveals the structure of F-tractin, a widely used peptide for visualizing the actin cytoskeleton, and introduces an optimized version that minim
Are you doing cell #biology? Then, let me guess, you likely stained #actin!
Our @jcb.org article reveals the structure of the popular actin probe F-tractin in complex with F-actin and introduces an optimized version for better imaging! A thread! 👀👇
rupress.org/jcb/article/...
11.02.2025 17:38 —
👍 11
🔁 2
💬 1
📌 0
Accounting for electron-beam-induced warping of molecular nanocrystals in MicroED structure determination pubmed.ncbi.nlm.nih.gov/39927752/ #cryoEM
11.02.2025 23:30 —
👍 2
🔁 1
💬 0
📌 0

(BioRxiv All) CryoEM-enabled visual proteomics reveals de novo structures of oligomeric protein complexes from Azotobacter vinelandii: Single particle cryoelectron microscopy (cryoEM) and cryoelectron tomography (cryoET) are powerful methods for unveiling… http://dlvr.it/THnVMD #BioRxiv #MassSpecRSS
05.02.2025 04:03 —
👍 2
🔁 1
💬 0
📌 0
Complex Water Networks Visualized through 2.2-2.3 A Cryogenic Electron Microscopy of RNA pubmed.ncbi.nlm.nih.gov/39896454/ #cryoEM
05.02.2025 05:44 —
👍 2
🔁 1
💬 0
📌 0
- identifies crucial contacts; very useful for selecting mutagenesis targets
- fixes model geometry. In cryo-EM, stereochemistry is usually the best priors you have!
- helps predicting models of weak complexes that AlphaFold3 cannot
06.02.2025 10:07 —
👍 2
🔁 1
💬 1
📌 0
Our findings emphasize how we use PDB models. It is critical to understand that PDB models are just that, models. They do not fully explain the underlying experimental data, have nuances in how they are representing the data, and as we show, can vary in how well the underlying data is explained.
16.12.2024 15:14 —
👍 10
🔁 1
💬 0
📌 0
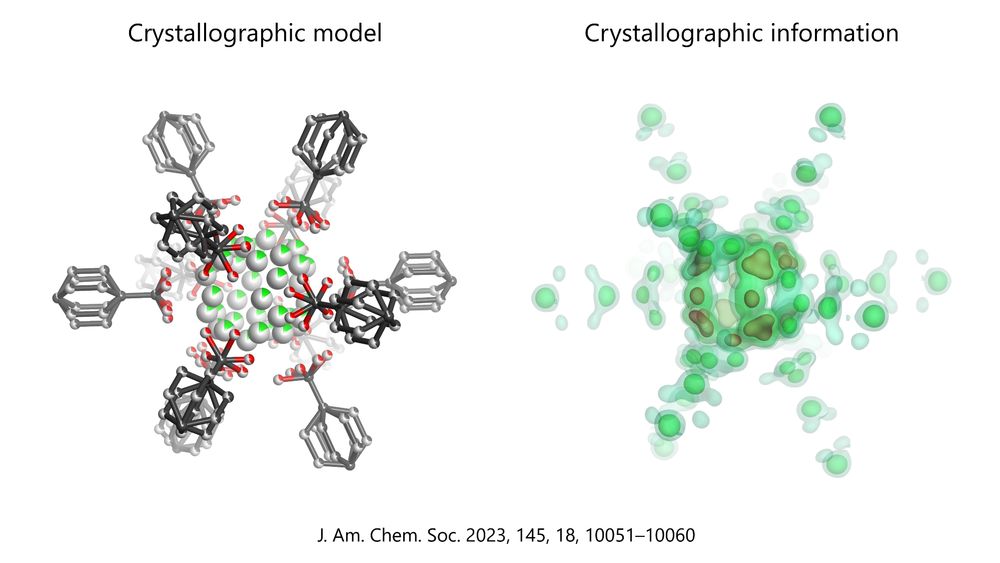
The difference between a model and the underlying crystallographic information is shown for the disordered MOF published in this paper: https://pubs.acs.org/doi/10.1021/jacs.2c13731. The model shows a ball&stick atomic structure, whereas the information (electron density calculated from structure factors) is displayed as density map with coloured contours.
Casual reminder that what many consider 'information' from #crystallography is actually a model. That is why structure factors must *always* be published, so that reviewers and anyone can see what lies behind the model. Without them, crystal structures are nothing more than 'pictures' in 3D.
30.11.2024 11:25 —
👍 29
🔁 2
💬 3
📌 1

a cartoon of homer simpson holding a card
ALT: a cartoon of homer simpson holding a card
Following a suggestion from @benecal.bsky.social I will refer to these threads as "skeetorials" and will use this gif logo.
Today's skeetorial is about the NIH Director's New Innovator Award with three different heroes (all in government).
1/n
23.11.2024 13:05 —
👍 133
🔁 41
💬 10
📌 16
Who are some amazing cryoEM/cryoET, mass spec, and light microscopy scientists coming to San Diego for ASCB? I’m making a list for a potential pre-ASCB workshop at UCSD December 13th. Spread the word and let me know if you know a good candidate to speak at such an event!
23.11.2024 17:55 —
👍 6
🔁 6
💬 0
📌 0
Here is a CryoEM starter pack I try to assemble
go.bsky.app/JmQksC
23.11.2024 18:43 —
👍 5
🔁 2
💬 1
📌 0
🚨 Our story on the TSC:WIPI3 lysosomal-recruitment complex is now live in Science Advances.
Equal-first co-authors @luptoncj.bsky.social & Laura D'Andrea. Work lead by @drellisdon.bsky.social. Funding from @deptofdefense.bsky.social & Australian Research Council.
www.science.org/doi/10.1126/...
20.11.2024 21:48 —
👍 58
🔁 19
💬 5
📌 0
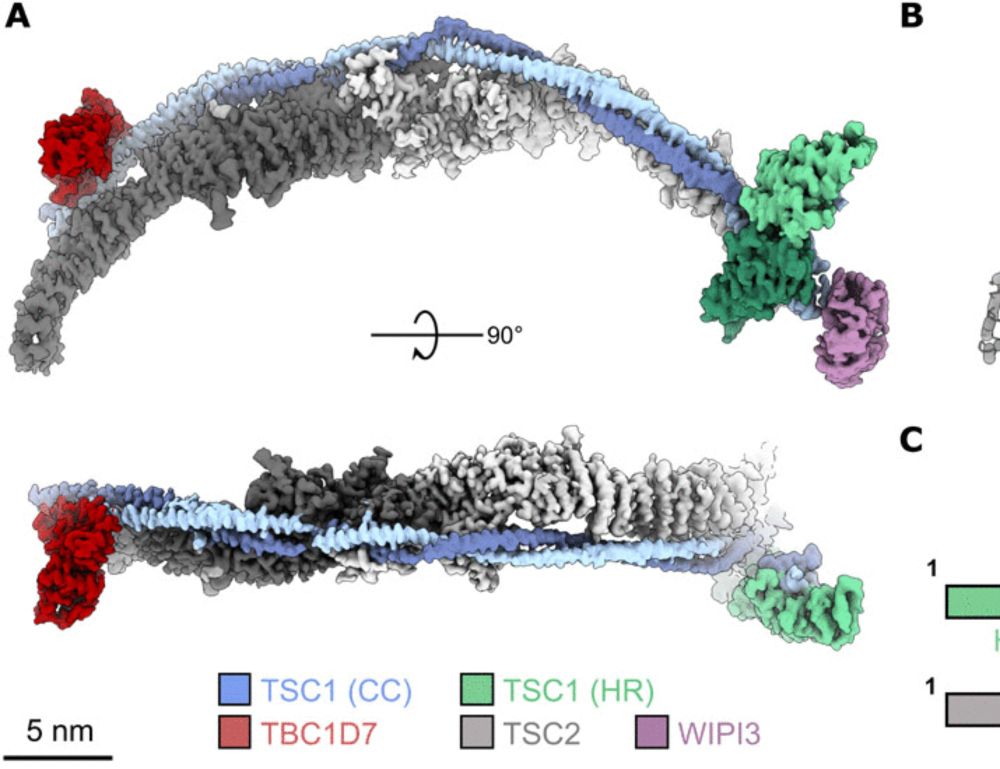
Structure of the human TSC:WIPI3 lysosomal recruitment complex
The complete TSC structure reveals how lysosomal targeting proteins and phosphoinositides coordinate mTORC1 inhibition.
Great to see our work on the #cryoem structure of the human TSC:WIPI3 lysosomal recruitment complex! Awesome effort from co-authors @charlesbj.bsky.social and Laura D'Andrea with @drellisdon.bsky.social doing the heavy lifting leading the project.
www.science.org/doi/10.1126/...
21.11.2024 00:35 —
👍 27
🔁 7
💬 0
📌 0

Cryo-electron micrograph image (or more properly, the average of 1,236 such images) of a 6-pointed star-shaped protein complex found in the interior of slime mold cells.
Question for the #CryoEM crowd: does anyone happen to recognize this lovely ~10-15nm diameter protein complex we stumbled across while imaging slime mold protein lysate (as one does)?
21.11.2024 03:30 —
👍 34
🔁 11
💬 5
📌 0