Here are the success rates of de novo pipelines based on which designs I could actually identify the methods for.
22.01.2026 15:47 — 👍 23 🔁 12 💬 2 📌 1

Mirdita Lab - Laboratory for Computational Biology & Molecular Machine Learning
Mirdita Lab builds scalable bioinformatics methods.
My time in @martinsteinegger.bsky.social's group is ending, but I’m staying in Korea to build a lab at Sungkyunkwan University School of Medicine. If you or someone you know is interested in molecular machine learning and open-source bioinformatics, please reach out. I am hiring!
mirdita.org
20.01.2026 11:07 — 👍 104 🔁 54 💬 7 📌 1
Second preprint of the year in which @sarahgersing.bsky.social from @rhp-lab.bsky.social mapped the effects of >7500 variants in glucokinase (GCK) on the interaction with the glucokinase regulatory protein so that we now have a 3D GCK scan (abundance, interaction, activity)
doi.org/10.64898/202...
18.01.2026 18:14 — 👍 23 🔁 6 💬 0 📌 0
This one was quite the journey! The paper describing the #ChlamyDataset is finally out and on the cover of Mol Cell!
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
09.01.2026 19:14 — 👍 88 🔁 35 💬 2 📌 10
Repeat after me: GENE 👏 EXPRESSION 👏 PROFILING 👏 DOES 👏 NOT 👏 A 👏 VIRTUAL 👏 CELL 👏 MAKE 👏
26.12.2025 12:11 — 👍 79 🔁 16 💬 4 📌 0

A Xmas tree assembled from a plastic molecular set, decorated with 3D printed molecular models.

A 3D printed molecular model.
Xtals tree, 2026 edition.
One structure for each user with finalized structure from current year.
#crystallography #chemtree #chemchat #chemsky
20.12.2025 00:07 — 👍 15 🔁 3 💬 0 📌 0
Brand new preprint from my lab, showing that TnpB, the ancestor of Cas12, acts as a gene drive in plasmids! And it turns out in conjugative plasmids that it acts as a primitive anti-self defense system, providing a potential link between its transposon effect and becoming CRISPR!
20.11.2025 22:17 — 👍 63 🔁 28 💬 2 📌 1


New preprint! We measured temperature- and pH-induced aggregation for over 18,000 natural and de novo designed protein domains!
19.11.2025 21:16 — 👍 121 🔁 42 💬 4 📌 3
Tired of knowing what you are looking for in your tomograms?
Why not try bluesky-less Frosina's new self-supervised algorithm for semantic segmentation and particle picking for #teamtomo (includes a new denoiser without the need for odd/even tomos)
Code is on GitHub if you want to try it out
19.11.2025 14:04 — 👍 52 🔁 11 💬 0 📌 0

A few py2Dmol updates 🧬
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
19.11.2025 08:15 — 👍 103 🔁 31 💬 1 📌 0
#1 lesson I give to protein designers for wet lab purposes : Please design with a Trp and/or multiple Tyr. Please do not design with 10 cysteines.
18.11.2025 13:48 — 👍 19 🔁 4 💬 2 📌 0
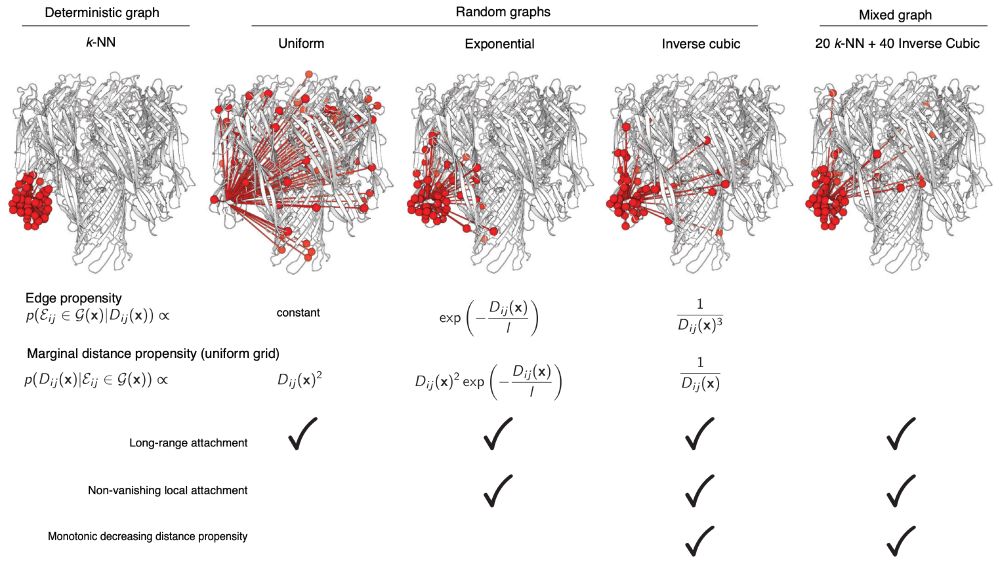
Has this idea of sampling neighbors from an inverse cubic distribution been used in protein structure GNNs at all since the idea was first presented in the Chroma paper ~3 years ago? I thought it was incredibly clever and expected it to catch on, but haven't seen it since
16.11.2025 21:01 — 👍 7 🔁 1 💬 2 📌 0

Efficient protein structure generation with sparse denoising models - Nature Machine Intelligence
A small and fast diffusion model is presented, which is able to efficiently generate long protein backbones.
We used a similar random neighbour scheme in salad (www.nature.com/articles/s42...), with 16 neighbours by residue index, 16 neighbours by distance and another 16 using the 1/d^3 scheme from Chroma, which worked reasonably well for protein backbone generation.
16.11.2025 21:18 — 👍 2 🔁 0 💬 1 📌 0
hey bsky! excited to share what i've been working on for a few years now.
cryoJAX: A Cryo-EM Image Simulation Library In JAX
www.biorxiv.org/content/10.1...
cryoJAX is a modular framework for implementing forward models of cryo-EM image formation. these can be used to build powerful data analyses!
14.11.2025 17:33 — 👍 13 🔁 2 💬 1 📌 2
Global Analysis of Aggregation Determinants in Small Protein Domains https://www.biorxiv.org/content/10.1101/2025.11.11.687847v1
13.11.2025 01:47 — 👍 11 🔁 12 💬 0 📌 0
Are you an early-stage graduate student (2nd or 3rd year) or early-stage postdoc based in the US or Canada, working primarily in Drosophila? Would you like to help improve the experience of all trainees working in Drosophila research? If so, read on.
(Please repost to reach a broad audience.)
12.11.2025 04:49 — 👍 87 🔁 182 💬 1 📌 2

a man with glasses and a mustache is wearing a white shirt and says yes .
ALT: a man with glasses and a mustache is wearing a white shirt and says yes .
#proteindesign #proteinbinder
100 designs
⬇️
34 got through the screening (ipTM, PAE, RMSD)
⬇️
4 ordered (gene synthesis + codon optimization)
⬇️
3 expressed and soluble
⬇️
1 interacts with its target !!!!
Now the story begins !
07.11.2025 12:03 — 👍 22 🔁 1 💬 2 📌 0

Can an AI tool help us better understand the origins of cancer?
Researchers from EMBL's Korbel Group have developed a new AI method – MAGIC – which, through a game of molecular laser tag, is shedding light on how chromosomal abnormalities form in cells.
www.embl.org/news/science...
29.10.2025 16:11 — 👍 13 🔁 5 💬 0 📌 0

Is 3D dragging you down? Wish you could instead use the 2D ColabFold representation for all your work? 🤓
Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)
29.10.2025 01:39 — 👍 29 🔁 4 💬 1 📌 1
New review on experimental datasets that can be used to benchmark protein force fields. And if that doesn’t tickle your fancy, the data can also be used to benchmark machine learning models for biomolecular structure and dynamics.
28.10.2025 15:51 — 👍 21 🔁 5 💬 0 📌 0

Icecream: High-Fidelity Equivariant Cryo-Electron Tomography
Cryo-electron tomography (cryo-ET) visualizes 3D cellular architecture in near-native states. Recent deep-learning methods (CryoCARE, IsoNet, DeepDeWedge, CryoLithe) improve denoising and artifact cor...
Our colleagues Vinith Kishore and Valentin Debarnot from the @ivandokmanic.bsky.social lab have come up with an amazing deep learning tool for denoising and filling the missing wedge in #cryoET data. I'm pleased to introduce Icecream🍧
23.10.2025 11:12 — 👍 53 🔁 20 💬 3 📌 5
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
22.10.2025 14:55 — 👍 14 🔁 18 💬 0 📌 0

Parents: Be sure check your children's Halloween candy carefully! Last year we found a restriction enzyme from New England Biolabs with a Dec 2003 expiration date inside a Snickers bar
22.10.2025 05:37 — 👍 61 🔁 9 💬 4 📌 1

Figure 1 from the preprint
Check out our preprint! With new molecular mechanisms, 140 subtomogram averages, and ~600 annotated cells under different conditions, we @embl.org were able to describe bacterial populations with in-cell #cryoET. And there’s a surprise at the end 🕵️
www.biorxiv.org/content/10.1...
#teamtomo
15.10.2025 06:26 — 👍 114 🔁 35 💬 8 📌 4

ProteinDJ logo
I am thrilled to release ProteinDJ: a high-performance and modular protein design pipeline. Our open-source workflow incorporates #RFdiffusion, #ProteinMPNN, #FAMPNN, #AlphaFold2 and #Boltz-2. It is a fast, free, and fun way to design proteins (1/5)
doi.org/10.1101/2025.09.24.678028 #proteindesign
28.09.2025 21:16 — 👍 88 🔁 31 💬 2 📌 6
Viro3D
🚨 New Web Resource Alert! 🚨 We're delighted to share Viro3D a database of >85000 viral protein structure predictions from >4400 human & animal viruses.
🔗 viro3d.cvr.gla.ac.uk
📄 www.embopress.org/doi/full/10....
@molsystbiol.org @cvrinfo.bsky.social @uofgmvls.bsky.social #Virology #AlphaFold 🧪 🦠
26.09.2025 11:36 — 👍 39 🔁 20 💬 1 📌 0
optimization, inverse problems, also proteins. ml at escalante. formerly: atomicai, xgenomes, broad, berkeley.
Professor of Bioinformatics. Executive Editor of the Database Issue at @narjournal.bsky.social. Proud Dad. Views my own.
🧬 Research group at EMBL Heidelberg, exploring genomes in motion | from SVs to disease mechanisms
Run by lab members | posting about science & lab life
Solve Biology
Head of Generative Genomics, Wellcome Sanger Institute, Cambridge, UK
Systems + Synthetic Biology, CRG, Barcelona
http://barcelonacollaboratorium.com http://allox.bio https://www.sanger.ac.uk/programme/generative-and-synthetic-genomics/
Genomics, Bioinformatics.
github.com/tobiasrausch
RNA & Metabolism. Ancient companions for Life. RNA as spatial organizer of the Cell. IDPs, biomolecular condensates and RNA fun @HymanLab. MBL Physiology '25. And birds.
ebird.org/profile/MjcyMDU0Nw
inaturalist.org/people/9261834
We are the largest biomedical science hub in the south of Europe, by the beach in Barcelona.
Recerca biomèdica amb vistes |
@researchmar.bsky.social @crg.eu @melisupf.bsky.social @embl.org @isglobal.org @ibe-barcelona.bsky.social @fpmaragall.bsky.social
computational biologist. all things genomic. working on cfDNA, cancer genomics, early detection, prenatal testing, epigenetics, transposable elements. building reproducible computational pipelines. seeking academic partners for collaboration.
DL for protein design & dynamics • stanford phd
gelnesr.github.io
Phage Biology, Bacterial defense systems, CRISPR-Cas, Microbial communities, Mucus interactions
http://www.fnobregalab.org
http://www.klebphacol.org
http://www.phage-collection.org
The Höfer lab studies the epitranscriptomic mechanisms of gene regulation based on NAD-capped RNAs in bacteria and bacteriophages. We are located at the Max Planck Institute for terrestrial Microbiology, Marburg, Germany and area Member of SYNMIKRO
MD, PhD. Postdoc in Slack and Sunagawa labs, ETH Institute of Microbiology. Typas lab alumna.
Bacteria as friends and foes.
PhD Student @ Schwille Lab, MPIB. Interested in the synthesis of protein design and synthetic cells. Eventually aiming to create life from scratch.
PhD student interested in molecular arms race and protein biology @cam.ac.uk.
MD, Russian National Research Medical University.
PhD student @Schwille Lab in Munich. Protein Design for synthetic cells!
Postdoc at UCL with James Reading. Previously at EMBL working with Wolfgang Huber. Biostats, R, cancer immunology
Biology-inspired physics | Information processing | PhD student in the Erzberger group at EMBL | she/her
The European Molecular Biology Laboratory drives visionary basic research and technology development in the life sciences. www.embl.org
Systems Microbiology @EMBL, combining high-throughput approaches & mechanism in bacteria, antibiotics, microbiome, infection, species diversity, cell envelope
https://www.embl.org/groups/typas/


















