Finally, thanks to the team! A huge shoutout to my friend and mentee @krithik-bs.bsky.social. Also infinitely grateful for #AlbertGu for his advice, and #MichaelMitzenmacher @pardissabeti.bsky.social for their mentorship and leadership. So much laughter while making this paper, can't wait for more.
21.03.2025 21:16 — 👍 1 🔁 0 💬 0 📌 0
This work shows that principled mathematical insights, like approximation of epistatic interactions, can provide an accessible and performant alternative to large foundation models—suggesting broader applicability beyond biological sequences.
21.03.2025 21:16 — 👍 2 🔁 0 💬 1 📌 0
We are excited about Lyra's potential to accelerate discoveries in molecular biology, therapeutic development, and protein engineering.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
Lyra makes cutting-edge biological modeling accessible to labs without extensive compute resources. Instead of relying on massive GPU clusters, Lyra empowers researchers to train state-of-the-art models directly on their own laptops.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
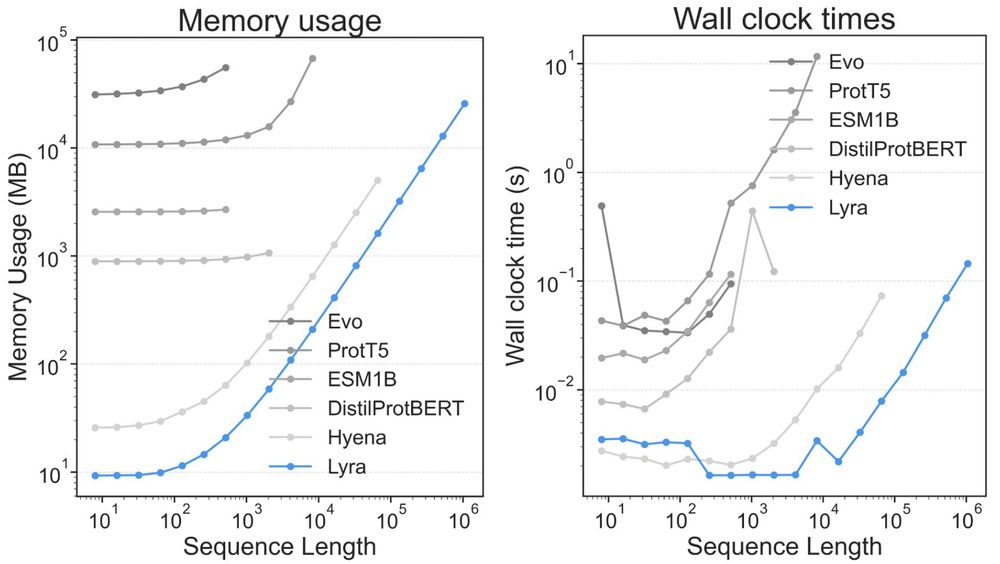
Lyra’s subquadratic O(N log N) complexity dramatically reduces memory (125x–2600x less than Evo and ESM-1b) and accelerates inference—up to 239x faster than ESM-1b, processing sequences up to 1M length.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
RNA-dependent RNA polymerases (RDRPs) are essential markers for RNA virus detection. Lyra achieves a near-perfect 0.998 true positive rate, matching LucaProt-ESM with over 60,000x fewer parameters, accelerating pathogen discovery without needing large-scale GPU infrastructure.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
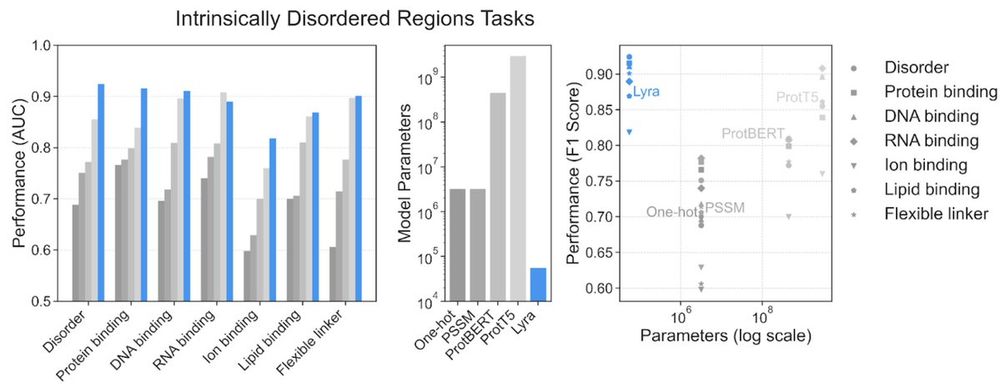
Lyra achieves SOTA results in 6 out of 7 intrinsically disordered protein region tasks, with an average AUC of 0.89, outperforming a ProtT5-based model (avg AUC 0.86). Lyra accomplishes this using only 55K parameters, compared to ProtT5’s 3 billion parameters—a >50,000-fold reduction in model size.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
Lyra’s consistently strong performance across different tasks using orders of magnitude fewer parameters allows researchers to spend less time optimizing models and more time generating biological insights.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
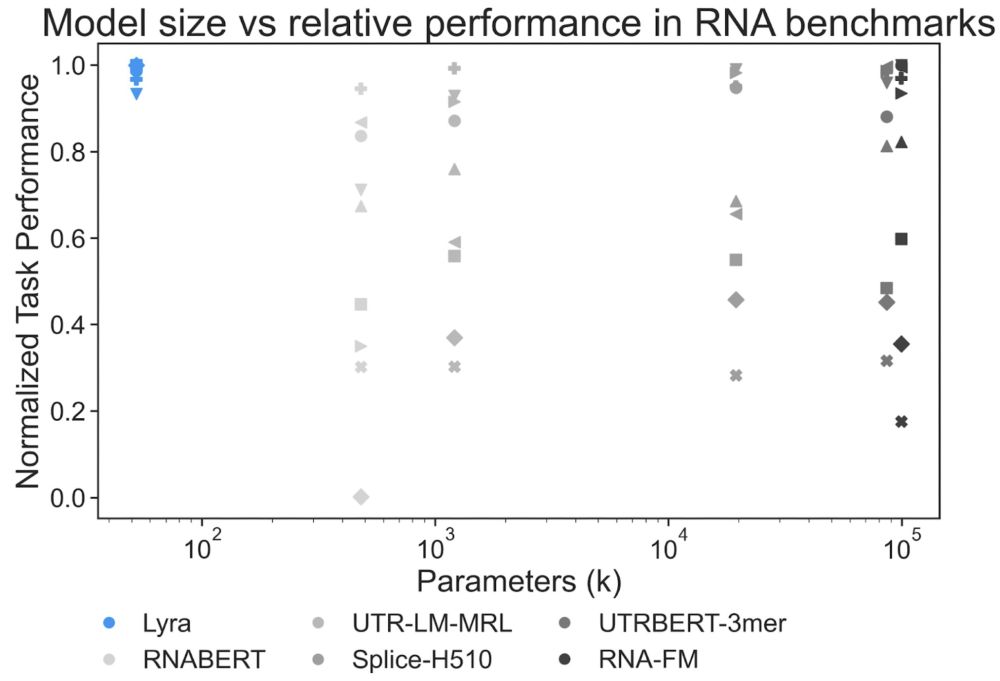
Lyra sets records in 5/9 RNA BEACON benchmarking tasks tested, including nearly solving the splice-site prediction dataset (98.89% accuracy vs previous best 50.55%) and almost doubling performance on structural score imputation (0.73 vs 0.42).
21.03.2025 21:16 — 👍 2 🔁 0 💬 1 📌 0
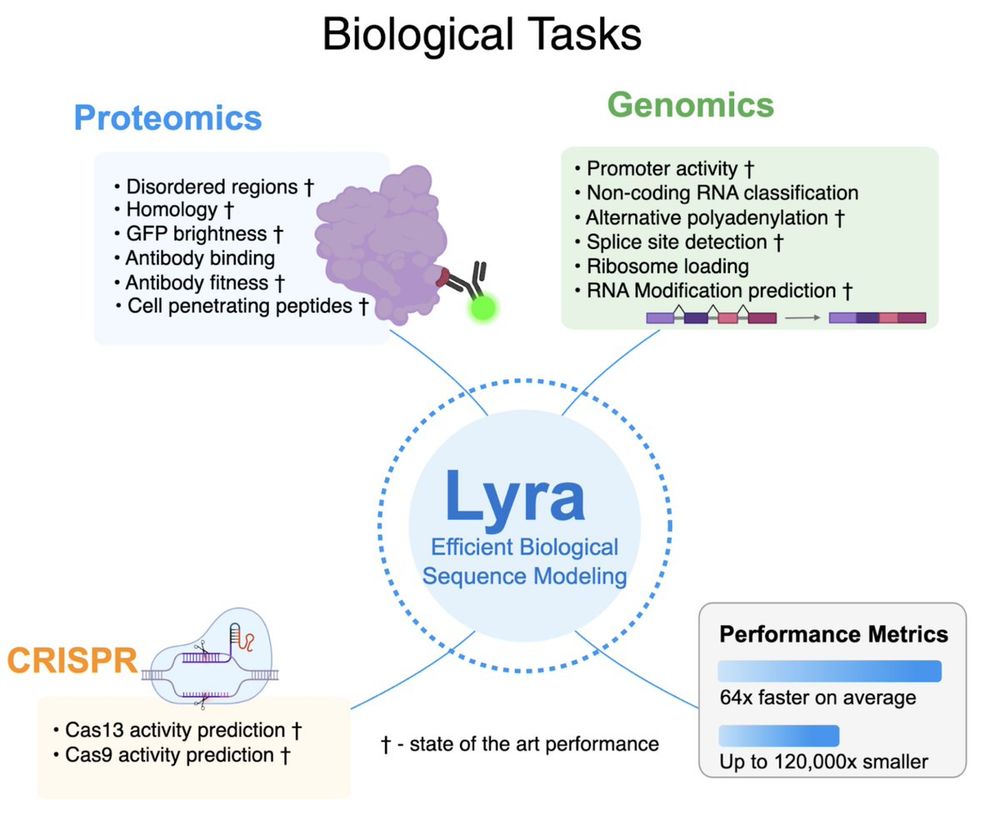
We tested Lyra on 101 diverse biological tasks spanning:
1. Proteomics
2. Genomics
3. CRISPR guide efficacy
Lyra set new performance records in 79 out of 101 tasks, w/ substantially smaller models than competing architectures.
21.03.2025 21:16 — 👍 2 🔁 0 💬 1 📌 0
We designed Lyra with two simple components: Projected Gated Convolutions (PGC), which enhance local feature extraction, and diagonalized State Space Models (S4D), which capture global epistatic interactions. In doing so, Lyra efficiently captures both global and local epistatic relationships.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
We drew a mathematical connection between State Space Models (SSMs) and polynomial approximation, showing how their hidden states can naturally approximate the polynomial terms that govern epistatic relationships. This makes SSMs ideal for modeling biological functions as multilinear polynomials.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
This perspective provides a principled mathematical framework for modeling sequence-function relationships.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
To unify biological sequence modeling across DNA, RNA, and proteins into a single computational framework, we revisited epistasis—the phenomenon where mutations influence each other—which can be characterized by multilinear polynomials.
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
Breaking down how biological sequences encode molecular functions remains a central challenge in computational biology. For example, given a GFP sequence, can we predict its fluorescence brightness?
21.03.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0

🧬 Meet Lyra, a new paradigm for accessible, powerful modeling of biological sequences. Lyra is a lightweight SSM achieving SOTA performance across DNA, RNA, and protein tasks—yet up to 120,000x smaller than foundation models (ESM, Evo). Bonus: you can train it on your Mac.
arxiv.org/abs/2503.16351
21.03.2025 21:16 — 👍 20 🔁 12 💬 1 📌 2
Entrepreneur
Costplusdrugs.com
Harvard/MIT MD-PhD student. Passionate about molecular evolution and translational genomics!
Scientist @broadinstitute.org researching at the intersection of pathogens, diagnostics, and public health. 🦠🧬🔬
Damon Runyon postdoctoral fellow in the Rice laboratory @ Rockefeller University | systems virologist | PhD Sabeti lab @ Harvard Virology | she/her/hers
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
PhD @Stanford @HazyResearch in AI Systems, Incoming Assistant Professor @Caltech CMS
CEO of FutureHouse, building an AI Scientist
Director of the Office of Biological Technologies of DARPA
I use genomics to study the evolution and spread of human pathogens and lead pathogen genomic analytics at the Broad Institute.
Also: @dpark@mstdn.science
ORCID: 0000-0001-7226-7781
GitHub: dpark01
Postdoc in the Sabeti Lab. Interested in the genomic epidemiology, evolution, and pathogenicity of RNA viruses 🦠🏳️🌈
Postdoc at @shaleklab & @sabeti_lab | Single-cell and infectious diseases | Ph.D. @embl @alexandrovteam | Alumni @Uniandes @cemop_lab | He/Him/His | 🇨🇴
Proteins, evolutionary models, unsupervised learning. Prev: Research Scientist MetaAI, PhD Berkeley. he/him.
MIT PhD Student - ML for biomolecules - https://hannes-stark.com/
AI for biology | Stanford and Arc Institute
MLing biomolecules en route to structural systems biology. Asst Prof of Systems Biology @Columbia. Prev. @Harvard SysBio; @Stanford Genetics, Stats.
Cofounder/CEO Octant
BoD Ginkgo Bioworks
Defense Science Board for Emerging Biotech
Fmr: Associate Professor, UCLA





