I’m incredibly proud of the progress we’ve made. What began as a vision to better understand how to engineer therapeutic enzymes is now taking shape through promising early data, a world-class scientific team, and a clear development roadmap.
16.06.2025 20:20 —
👍 0
🔁 0
💬 0
📌 0
Announcing the launch of our #Seed #Funding #Round as we take the next step in our mission to transform the future of #enzyme #replacement #therapies for #lysosomal #storage #diseases.
With a passionate team, early proof-of-concept data, and a clear path to the clinic, we’re ready to scale!
16.06.2025 20:10 —
👍 0
🔁 1
💬 0
📌 1
Let’s see if I can make it next year!
20.05.2025 17:43 —
👍 2
🔁 0
💬 0
📌 0
I won’t attend but please get in touch with Susan and Alice if you want to know more about our #advanced #therapeutics for #lysosomal #storage #disorders.
25.04.2025 18:23 —
👍 2
🔁 0
💬 0
📌 0
Had a great time the past 2 days hosting my good friend and colleague Dr Boyd McKew from @universityofessex.bsky.social obviously lots of good chat about sequencing, synthetic biology, diving and beers :)
25.04.2025 18:19 —
👍 0
🔁 0
💬 0
📌 0
Pushing the boundaries of therapeutics here at @edinburgh-uni.bsky.social If you want to know more about our work in the hub on #Fabry and #MPSIVA get in touch. #ML #engbio #rare #diseases
10.04.2025 19:02 —
👍 0
🔁 0
💬 0
📌 0

Latest addition to our #enzyme #replacement #therapy platform: the Getinge Applikon #miniBio 500mL bioreactor. Excited to start new pilot projects on up-scaling enzyme production.
Thanks Scottish Enterprise to continuously support ZYTHERA's journey in/outside the lab.
#rare #disease #bioproduction
26.03.2025 12:44 —
👍 0
🔁 0
💬 0
📌 0
It’s #rare #disease #day! There are ~10,000 rare diseases, most lacking a cure or treatment. The path that patients and carers go through is something no one should ever experience.
Let’s harness the #new #technologies from our labs to deliver personalised medicine, because nobody is rare!
28.02.2025 17:46 —
👍 0
🔁 0
💬 0
📌 0
Load <return>
Press play on tape
It never gets old :)
25.02.2025 16:36 —
👍 1
🔁 0
💬 0
📌 0
Some of our recent work:
- Temporal Dirichlet Variational Autoencoder: www.nature.com/articles/s41...
- Protein engineering by variational energy approximation
www.nature.com/articles/s41...
24.02.2025 20:24 —
👍 1
🔁 0
💬 0
📌 0
Welcome to the Stracquadanio Lab
A data-driven synthetic biology lab using machine learning and synthetic biology to understand cancer biology.
Hi there! Officially starting my new academic account on @bsky.app.
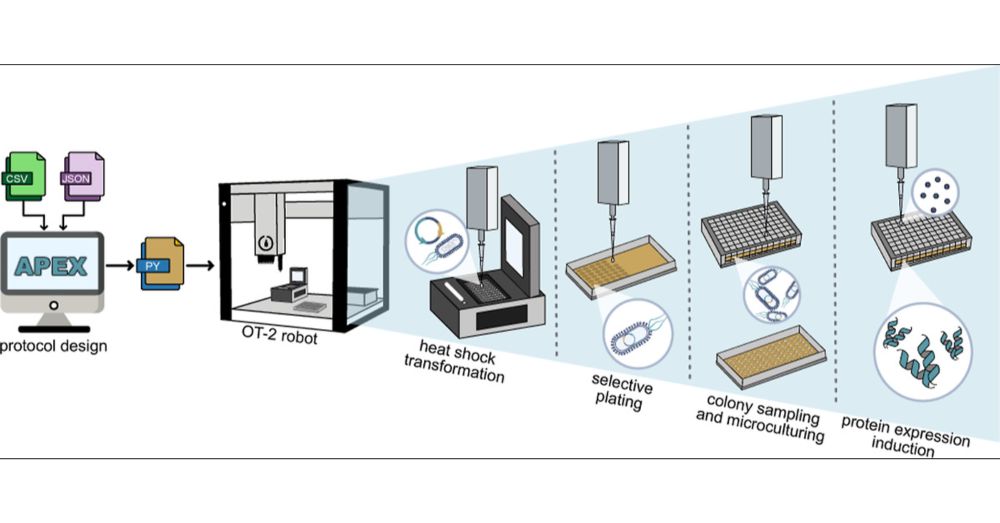
My group works on generative #AI models and #hightroughput #protein #expression and #screening platforms to engineer potent therapies for #lysosomal #storage #diseases. Learn more at: www.stracquadaniolab.org
24.02.2025 20:20 —
👍 3
🔁 0
💬 1
📌 0