Unless I misunderstood and you are asking about spatially proximal genes in general. In which case it could either, i.e. regulated (for example same TAD) or incidental.
16.01.2026 20:21 — 👍 1 🔁 0 💬 1 📌 0
These are sister chromatids (copies of the same chromosome after replication). They are held together by cohesin and other proteins complexes so the proximity is not accidental. if you image with enough temporal and spatial resolution you can incidentally see them separate some times.
16.01.2026 20:18 — 👍 2 🔁 0 💬 1 📌 0
One big take away (that took some while for us to reconcile here) is to not assume when you see a bright focus in a cell that a non-stochiometric phenomenon is occurring.
16.01.2026 14:12 — 👍 5 🔁 0 💬 1 📌 0
4) It is possible that the clusters we observe contain multiple genes that are firing in sync, but our simulations suggest that this is likely rare and the fact that we don’t see thousands of “clusters” is due to differences in Pol II occupancy at genes and technological limitations.
16.01.2026 14:12 — 👍 3 🔁 0 💬 1 📌 0

3) One observation that was quite convincing to us, is that when we look at the same locus firing on sister chromatids, when they transiently separate, we clearly see two Pol II clusters. By cross linking capture methods these would show a high contact frequency, and by static FISH high overlap.
16.01.2026 14:12 — 👍 5 🔁 0 💬 1 📌 0
2) When looking at fixed cell images with diffraction limited resolution, there might be multiple clusters at each FISH spot which are not resolvable.
16.01.2026 14:12 — 👍 3 🔁 0 💬 1 📌 0
A few points: 1) When interpreting evidence from chromatin conformation methods it is important to keep in mind that live imaging work has now shown that these contacts tend to be quite transient and infrequent in live cells.
16.01.2026 14:12 — 👍 2 🔁 0 💬 1 📌 0
Great analogy! Thanks
15.01.2026 20:11 — 👍 0 🔁 0 💬 0 📌 0

HLA-Shuttle: A system for enhancing antigen presentation in immunologically cold tumors
HLA-Shuttle enhances production of HLA class I molecules in neuroblastoma, leading to the discovery of tumor-associated antigens.
We are happy to have contributed single-molecule tracking and super-resolution imaging to this great work from Nik Sgourakis' lab. HLA-Shuttle restores antigen presentation in immunologically cold tumors enabling the identification of new targets for immunotherapy.
www.science.org/doi/10.1126/...
05.01.2026 19:34 — 👍 4 🔁 0 💬 0 📌 0
Huge props to our grad student @manyakapoor.bsky.social for developing the simulations and the new MS2 analysis amongst other things! We also thank the transcription community for their invaluable feedback and always welcome further discussion😊
18.09.2025 15:24 — 👍 3 🔁 2 💬 0 📌 0

WE ARE HIRING! The lab got the Packard AND an R35! Please spread this PostDoc Ad around! Philly and Upenn are a great place to live and work!
13.05.2025 19:28 — 👍 13 🔁 7 💬 2 📌 0
5/n: Please reach out directly to me if you are interested. This is a unique opportunity at a time where increased centralized resources are critical to maintain and accelerate the pace of scientific discovery.
11.04.2025 10:48 — 👍 1 🔁 0 💬 0 📌 0
4/n We are seeking a microscope specialist/engineer. The role will include developing and implementing new microscope hardware, control software, customized analysis pipelines as well as collaborating with core users.
11.04.2025 10:48 — 👍 0 🔁 0 💬 1 📌 0
Advanced Core for Microscope Engineering (ACME) | Advanced Core for Microscope Engineering | Perelman School of Medicine at the University of Pennsylvania
3/n We will provide custom built ad hoc optical microscopy solutions, advanced image analysis, and consultation services to researchers across campus and beyond. We will implement technology in response to experimental needs instead of working within the bounds of commercially available technology.
11.04.2025 10:48 — 👍 0 🔁 0 💬 1 📌 0
2/n: ACME is co-directed by myself (Mustafa Mir), Melike Lakadamyali @melikel.bsky.social (www.lakadamyali-lab.com), Yihui Shen (www.imaging-systems-metabolism-lab.com), and Andrea Stout @aleestoutphd.bsky.social (www.med.upenn.edu/cdbmicroscopycore)
11.04.2025 10:48 — 👍 0 🔁 0 💬 1 📌 0

1/n We have an exciting new initiative at UPenn to provide custom advanced microscopy solutions: Advanced Core for Microscope Engineering (ACME): www.med.upenn.edu/cdbacme/ We are looking for a specialist to help run this new facility, job description is attached please help share widely!
11.04.2025 10:48 — 👍 10 🔁 9 💬 1 📌 1
9/ We welcome feedback, questions, and critical discussion from the community. Huge thanks to all our amazing co-authors for their insight, support, and collaboration throughout this work.
09.04.2025 14:31 — 👍 1 🔁 0 💬 0 📌 0

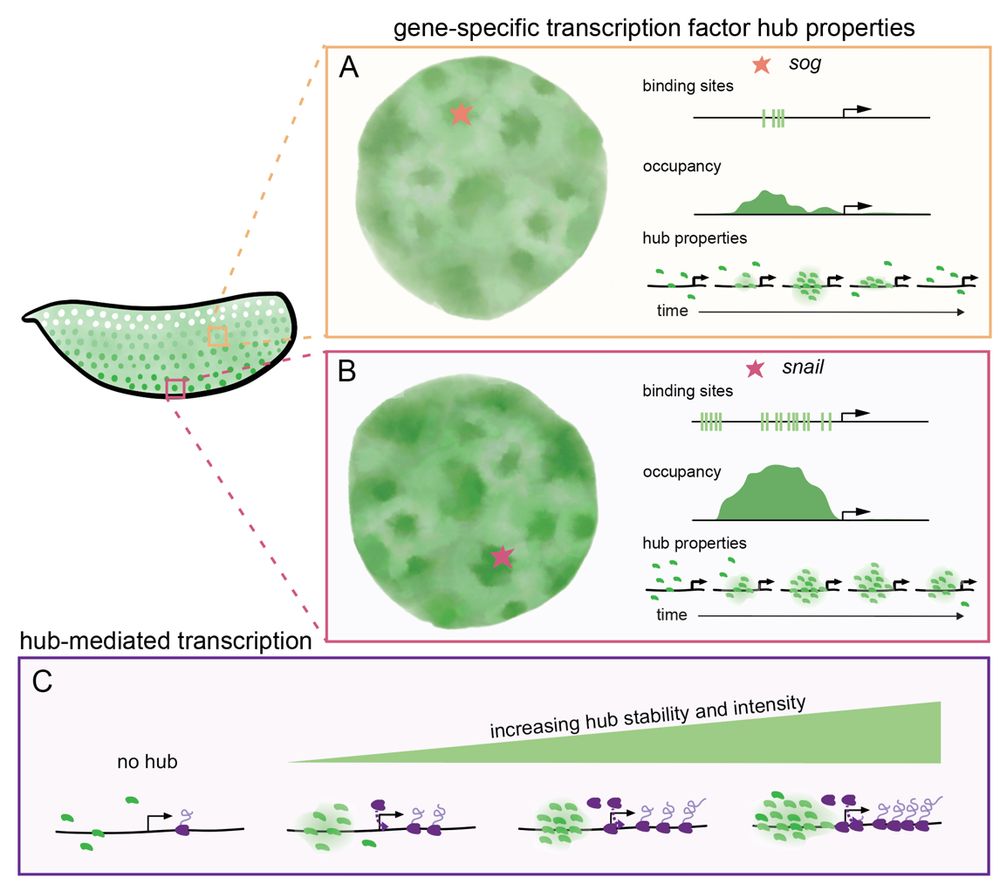
8/n: We suggest that enhancers encode the biophysical properties of transcription factor hubs (intensity and stability) through binding affinities and cofactors. Hub properties dictate gene occupancy and transcriptional kinetics, suggesting hubs are active executors of enhancer-encoded regulation.
09.04.2025 14:31 — 👍 1 🔁 0 💬 1 📌 0
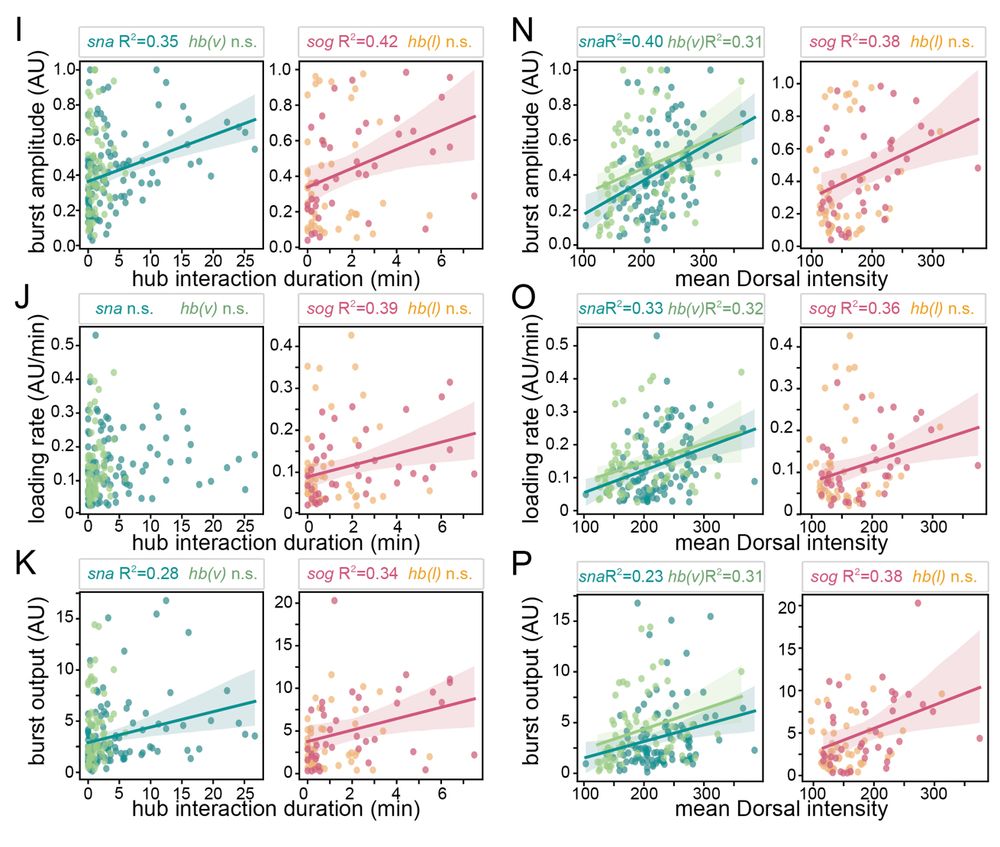
7/n: Hub intensity and duration correlate with burst amplitude, loading rate, and output suggesting that persistently interacting hubs with more molecules tune gene expression. Consistent with the idea that hubs facilitate transcription factor binding and promote expression.
09.04.2025 14:31 — 👍 2 🔁 0 💬 1 📌 0
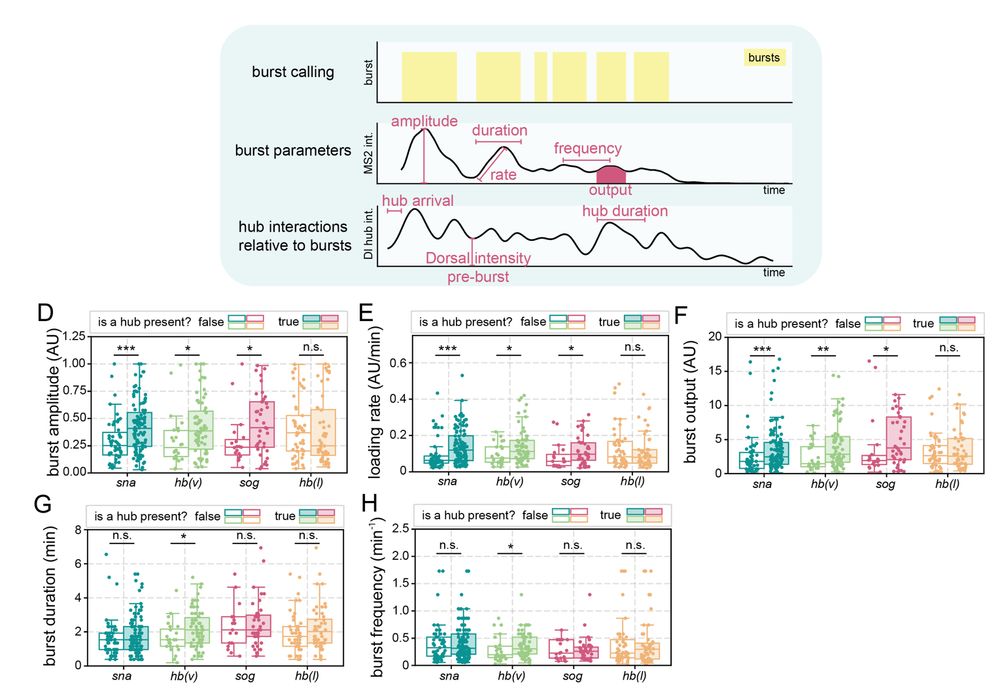
6/n: Hub presence at a gene is correlated with increased transcription burst amplitude, Pol II loading rate, and total output. Surprisingly, transient interactions between Dorsal hubs and hunchback (lacking Dorsal binding) boosts transcription in ventral nuclei where Dorsal concentration is highest
09.04.2025 14:31 — 👍 2 🔁 0 💬 1 📌 0
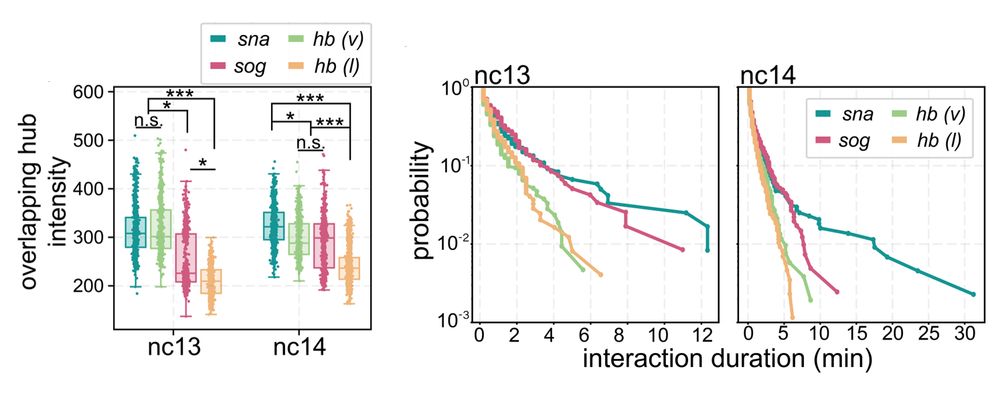
5/n: We found:
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
09.04.2025 14:31 — 👍 1 🔁 0 💬 1 📌 0
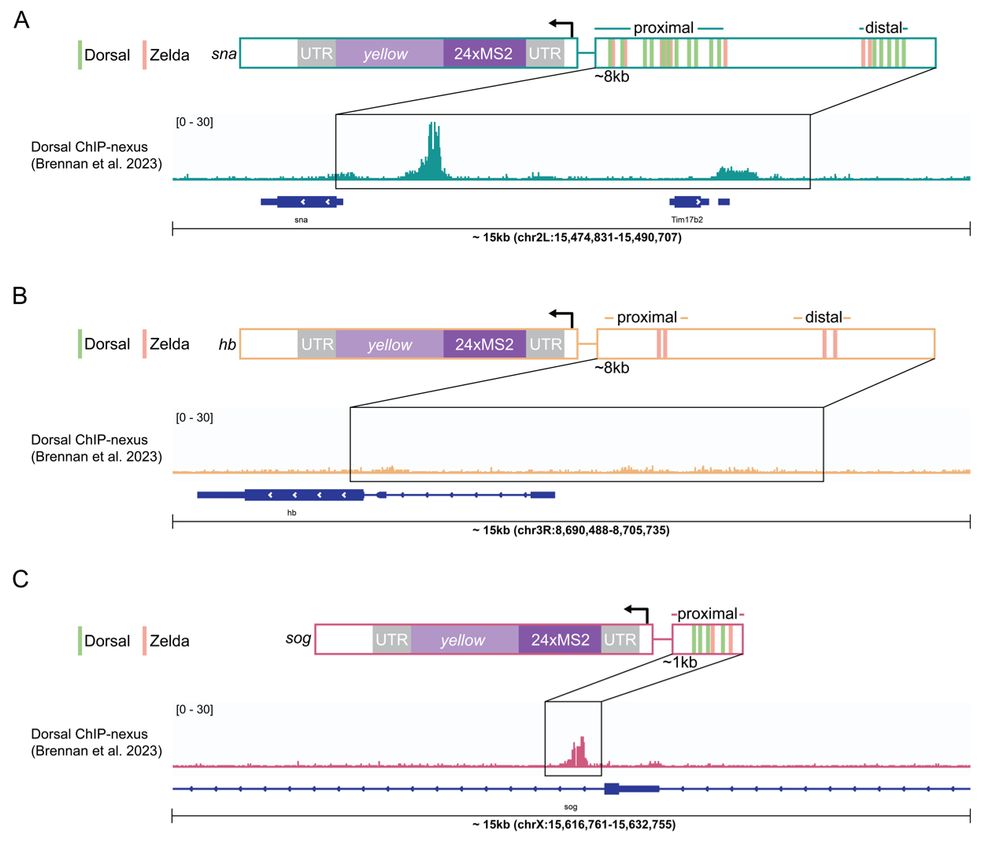
4/n: Next we examined Dorsal hub interactions at two target genes with distinct Dorsal occupancy and enhancer grammars, snai and sog, and as a negative control examined hunchback which has no dorsal sites.
09.04.2025 14:31 — 👍 1 🔁 0 💬 1 📌 0
3/n: First, we find that hubs form rapidly after mitosis and last through interphase. Nuclear hub density doesn't depend on nuclear concentration but hub intensity does, suggesting that hub formation may be independent of concentration, but hubs accumulate more protein with increased concentrations.
09.04.2025 14:31 — 👍 1 🔁 0 💬 1 📌 0
2/n: Transcription factor hubs (dense local accumulations) regulate gene expression by promoting the frequency of binding. They are often discussed as having uniform properties based on protein sequence and concentration. We wondered if their biophysical properties vary in a gene dependent manner.
09.04.2025 14:31 — 👍 4 🔁 0 💬 1 📌 0
In light of policies to defund biomedical research, and with judicial hearings happening today, we hope to highlight the many ways biomedical research improves people's lives, and to convey how damaging the proposed cuts would be... not just for us researchers, but for everyone.
21.02.2025 21:06 — 👍 34 🔁 17 💬 2 📌 0
And all the MS2 interaction analysis was done by an amazing bioengineering graduate student @manyakapoor.bsky.social who just joined Bsky!
12.02.2025 16:49 — 👍 1 🔁 0 💬 0 📌 0
Major oversight in my earlier postdoc: This work was led by our wonderful postdoc @mukherja.bsky.social who also built the microscope that enabled these experiments
12.02.2025 16:18 — 👍 1 🔁 0 💬 1 📌 0
We can't follow the same cluster but see a shift in cluster kinetics across nuclei. We can't say anything about initiation factors since we are just looking at pol2 and we are testing effects of recruitment vs elongation with the drugs. Will check our language to make sure this is clear. Thanks.
12.02.2025 13:56 — 👍 1 🔁 0 💬 0 📌 0
Forever developmental biologist at ❤️ studying the blood-brain barrier #BBB in the best model system #zebrafish to understand how different molecular and cellular interactions affect BBB function. www.obrownlab.com
Assistant Professor @UPenn.bsky.social
www.CarmanLiLab.org
Hereditary cancers, BRCA1/2, mouse models, organoids, single-cell omics
Enhancers, 3D genome organisation, pluripotent stem cells
Babraham Institute and Enhanc3D Genomics
3D genome, chromatin regulation, multi-omic TechDev @ New York Genome Center 🗽, via WEHI 🇦🇺
Structural biologist and biochemist. CNRS researcher at CBM Orléans @cbm-upr4301.bsky.social. Interested in protein modifications & interactions. Also husband, dad of 2, friend, ☧. Personal website: msuskiewicz.github.io
Biochemist fascinated by visualizing biological processes with the finest spatiotemporal detail; Understand the human immune system to guide novel approaches for combating viral infections and cancer; Postdoc at Cissé Lab / MPI-IE
Scientist/Immunologist. Immune Oncology/Tolerance. 3R.
Broad interest in science. My views are my own. Retweets, likes ≠ endorsement. 🇨🇭🌏
I really enjoy moving liquids around in a particular order
Will continue doing experiments until someone stops me
Searching for Utah jazz community on here
LMCB Group Leader at UCL. Cis-regulatory control of cell fate choice in development.
https://delaslab.com/
A Drosophila enthusiast with a strong interest in disease modeling
Assistant Professor @PennCDB @IRM_UPenn @PennIDOM @chronosleepinst, asking How circadian rhythms shape pancreatic islet development and diabetes. https://j-radlab.com
MD/PhD Student at the University of Pennsylvania
Derek Oldridge & Laura Vella Labs, Children's Hospital of Philadelphia
orcid.org/0000-0001-9799-8089
Interests - Germinal Center Biology | Human Immunology | Spatial & Single-Cell Omics | Really Good Bread
Presidential Assistant Professor of Physiology at the University of Pennsylvania.
Our laboratory in the MCB department at UC Berkeley studies how tissues, cells, and subcellular dynamics coordinate to generate organs and initiate physiologies. www.swinburnelab.org
Our mission: To foster inclusivity and equity in the Philadelphia
biomedical research community by showcasing cutting-edge research from scientists of all backgrounds and providing networking and career development opportunities.
Associate Professor at Duke | Director of Duke Spark | AI in Medical Imaging
Associate Professor 🧑🔬🧑🏫 at McGill University 🇨🇦 CRC in Spatial organization of living systems 💧🦠🪱🔬
https://weberlab.ca/
PhD student @ UPenn | Early mammalian development, mitosis, & microscopy











