Let's start the week with this movie kindly shared by @loxstoplox.bsky.social
The movie shows a mouse embryo labeled with Formin-2-GFP and SPY650-DNA during nuclear envelope breakdown. Note the nuclear dilution of Formin-2-GFP immediately before the SPY650-DNA labeled chromosomes start compaction.
23.06.2025 10:54 — 👍 8 🔁 3 💬 1 📌 0
Rewriting the rules of cell division
Microtubules get help from actin filaments during mitosis
New findings in Science demonstrate that successful mitosis may draw on more mechanisms than commonly assumed.
Learn more in this #SciencePerspective: scim.ag/4dFbqnp
30.05.2025 13:56 — 👍 52 🔁 10 💬 1 📌 0
Thank you Madhura!
28.05.2025 19:10 — 👍 1 🔁 0 💬 0 📌 0
Thank you so much Sam!
27.05.2025 02:29 — 👍 1 🔁 0 💬 0 📌 0
Actin sun.
25.05.2025 04:07 — 👍 152 🔁 22 💬 5 📌 2
🤣🤣🤣🤣
24.05.2025 20:07 — 👍 0 🔁 0 💬 0 📌 0
Thank you so much Pablo!
24.05.2025 18:38 — 👍 1 🔁 0 💬 0 📌 0
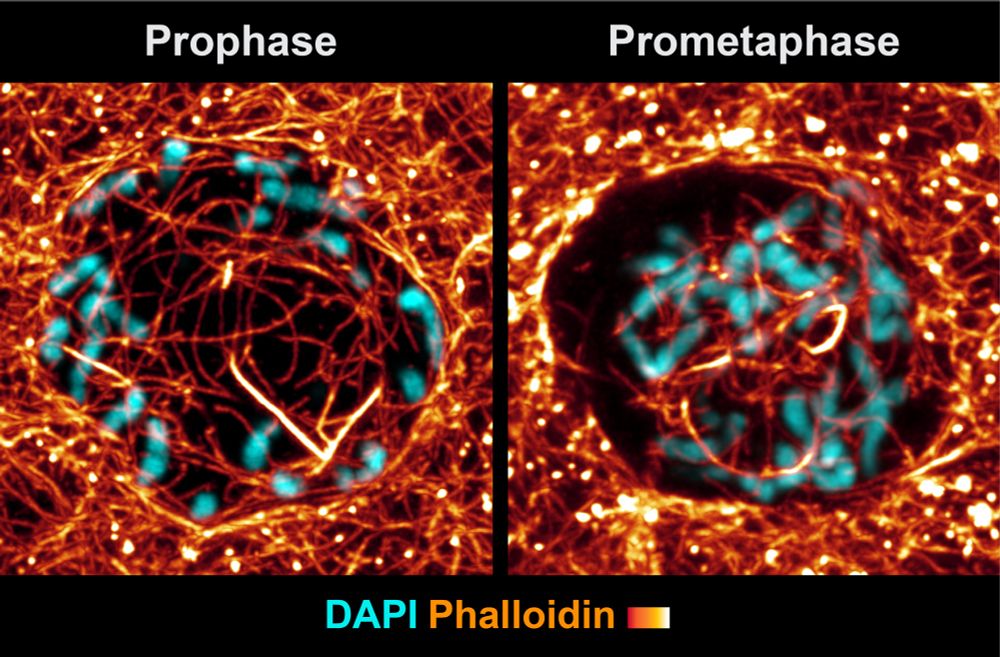
You appear to suggest that nuclear actin filaments disassemble during interphase. However, using phalloidin labeling we observed that nuclear actin density increases over the course of cell cycle progression.
24.05.2025 16:30 — 👍 0 🔁 0 💬 0 📌 0

Hi Robert, I’m assuming this is in response to Manuel’s post. I’m very familiar with your paper, nice work! Again, I disagree that the conclusions in my paper are similar to yours. In fact, here’s an example from your paper where we reach very different conclusions.
24.05.2025 16:30 — 👍 0 🔁 0 💬 1 📌 0
#FluorescenceFriday
23.05.2025 20:05 — 👍 5 🔁 0 💬 1 📌 0
A big thank you to our collaborators and the entire lab for their support inside and outside of lab! And huge shoutout to UPenn alum and actin guru @aaandmoore.bsky.social for key technical advice during the early stages of the project!!
23.05.2025 20:04 — 👍 5 🔁 0 💬 1 📌 0
I won’t spoil the details of the entire paper here, but we also identified a network of branched actin at the metaphase spindle periphery that modulates spindle size, offering a new explanation for the spindle scaling behavior seen in large acentriolar cells.
23.05.2025 20:02 — 👍 7 🔁 1 💬 1 📌 0
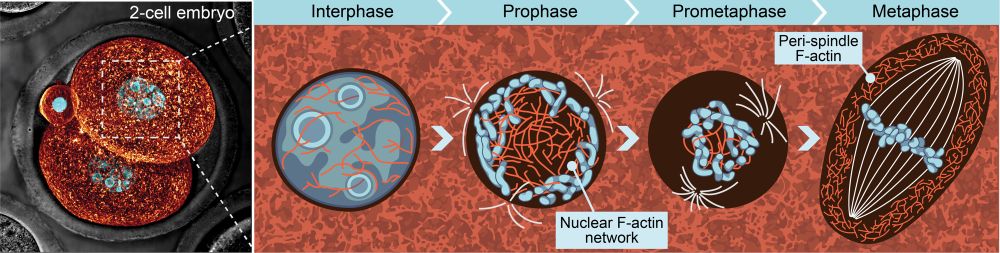
Surprisingly, network contraction is not driven by myosin-2. We instead found that contractile stress is generated by filament disassembly within the crosslinked network. We propose that after nebd, formin dilution from the nuclear region triggers filament disassembly and network contraction.
23.05.2025 20:01 — 👍 9 🔁 0 💬 1 📌 0
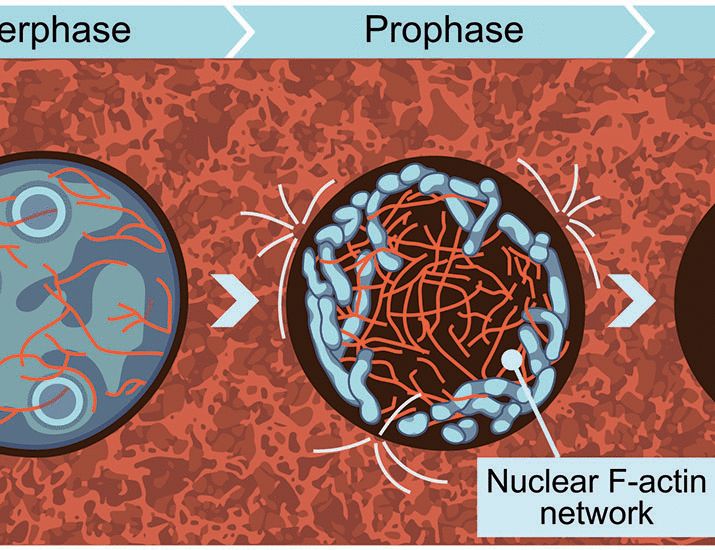
Actin to the rescue! A network of nuclear actin cables captures prophase chromosomes and contracts following nuclear envelope breakdown, gathering chromosomes towards the cell center. We propose that this mechanism of chromosome organization is required to achieve mitotic fidelity.
23.05.2025 19:59 — 👍 18 🔁 6 💬 1 📌 0
Even more puzzling, the first phase of mitotic chromosome organization occurs independently of spindle microtubules. So then what cellular component generates the necessary force to organize chromosomes?
23.05.2025 19:59 — 👍 3 🔁 0 💬 1 📌 0
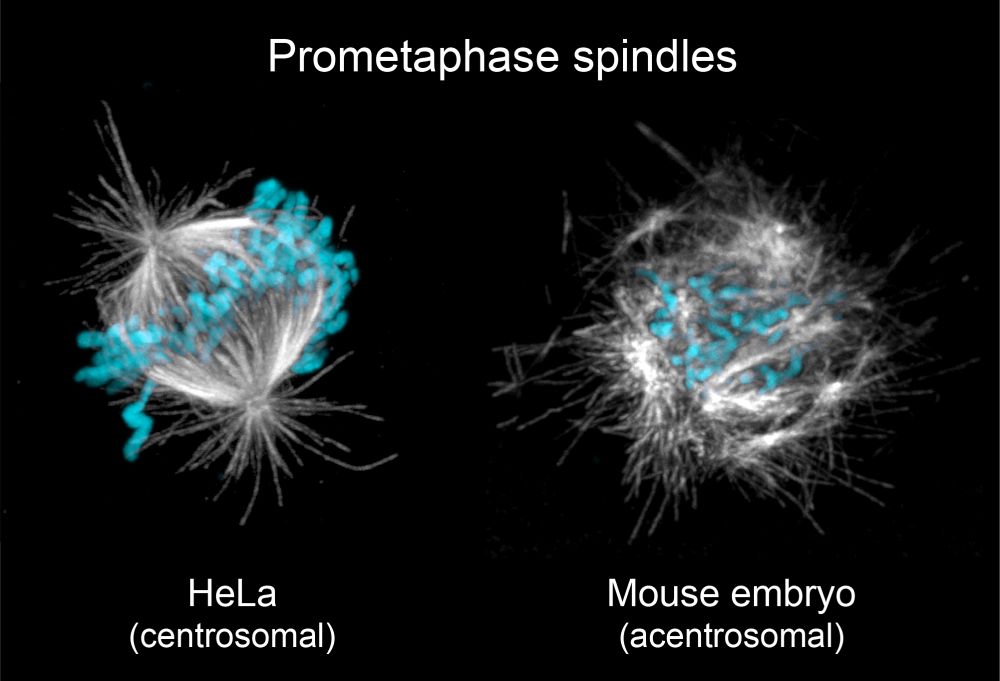
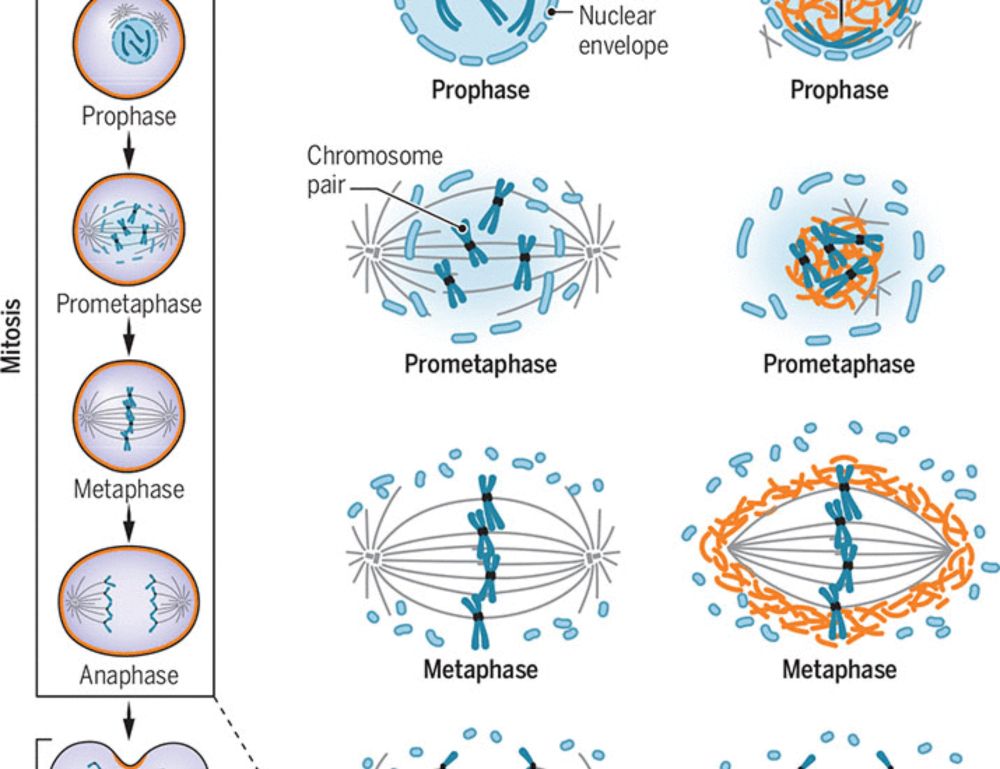
It’s therefore puzzling that spindle assembly in the early embryo is highly inefficient, exemplified by comically disordered spindles during early mitosis (shown below).
23.05.2025 19:58 — 👍 5 🔁 0 💬 2 📌 0
In the classical view of mitotic cell division, the spindle apparatus maintains principal control of chromosome capture and alignment. Any breakdown in spindle function can result in chromosome mis-segregation, producing daughter cells with abnormal chromosome number.
23.05.2025 19:57 — 👍 9 🔁 3 💬 1 📌 0
I’m excited to announce that my paper describing non-canonical mitotic mechanisms in the early mouse embryo is out in @science.org ! (link at end of 🧵)
23.05.2025 19:56 — 👍 190 🔁 44 💬 8 📌 1
The first part of the paper is indeed closely aligned with great work from the Lenart lab. It’s also important to note that our paper focuses on mitosis, whereas much of the work you referenced centers on meiosis.
23.05.2025 13:28 — 👍 3 🔁 0 💬 0 📌 0
Regarding your comments on novelty: while it’s true that our work builds on the results of others (including your own), I disagree with the comparison to great work from the Schuh, Mogessie, or Grosse labs.
23.05.2025 13:27 — 👍 1 🔁 0 💬 1 📌 0
Hi Manuel, big fan of your work! I’d like to point out that the manuscript you reviewed is not the version that was ultimately published. Once you’ve had a chance to read the final version, I’d really appreciate hearing your thoughts.
23.05.2025 13:25 — 👍 1 🔁 0 💬 1 📌 0
Thank you Christophe!! I’ll post a thread later today
23.05.2025 13:25 — 👍 1 🔁 0 💬 0 📌 0
Actin does it again #cytoskeletonwar
23.05.2025 10:02 — 👍 39 🔁 8 💬 2 📌 0
Quincy gets my vote for Off the Wall alone!
13.05.2025 23:15 — 👍 1 🔁 0 💬 0 📌 0
Dynamic mitochondria labeled with PKmito #FluorescenceFriday
25.04.2025 17:04 — 👍 20 🔁 2 💬 0 📌 0
Thank you!
12.04.2025 00:01 — 👍 1 🔁 0 💬 0 📌 0
DM1A never disappoints 🤩 #FluorescenceFriday
11.04.2025 23:32 — 👍 68 🔁 7 💬 2 📌 1
What started as a brief chat with @buvansr.bsky.social and @vgelfand.bsky.social at the 2023 CellBio meeting @ascbiology.bsky.social has grown into our published collaborative work on single vimentin IF dynamics and organization in cells. Love when stuff like that happens.
10.03.2025 18:39 — 👍 29 🔁 12 💬 1 📌 0
Beautiful! COS-7?
22.02.2025 05:50 — 👍 0 🔁 0 💬 0 📌 0
I am a Software Architect for AI Solutions, Python Fan, Microscopist Image Analyst and really like Skiing.
Science is like magic but real.
Postdoc in Elaine Fuchs Lab @The Rockefeller University. Previously, PhD student in Eduardo Villablanca Lab @Karolinska Institutet.
#stemcell-immuneinteraction #tissueimmunology
Mammalian cell biology and development lab at @rockefelleruniv.bsky.social & @hhmi.bsky.social ▪️Epithelia, stem cells, inflammation, cancer and more!
Deputy Editor Science magazine
Areas of responsibility include: Cell biology, cellular microbiology, membrane traffic, protein targeting, protein folding, organelle biogenesis, cytoskeleton, cell polarity, prion biology, developmental biology, stem cells
Junior Group Leader in Münster, Germany.
Morphogenesis, collective cell migration, cytoskeleton, cell-adhesion and self-organization/emergent behavior in #Drosophila.
Animal photos: 📷 instagram.com/maikscritters 🐸🐍
Assistant Professor @RiceBioE | PhD @PennBioengineering
women’s health • biomaterials • immune engineering
https://swingle-lab.com
Neuroscientist turned gene editor!
Postdoc in the Ahrens-Nicklas & Musunuru labs | Neuroscience PhD from Penn, Korb lab | Barnard College alum | she/her
Combining physical modeling, AI and computer vision to understand how cells self-organize into embryos and tissues. ERC DeepEmbryo. Team headed by @herveturlier.bsky.social
Asst. Prof. at UNIGE
Evolutionary cell biology & multicellular developmental diversity of protists.
www.dudinlab.com
#Ichthyosporea, #Multicellularity, #Evolution, #Embryo, #Development, #Protist, #UExM, #Cytoskeleton, #Actin, #Expansion #Microscopy
Cell & developmental biologist. Exploring instructive cues encoded in cell shape – in vascular development, morphogenesis and beyond. https://herbertlab.com/ @uniofmanchester.bsky.social @fbmh-uom.bsky.social @mcf-uom.bsky.social
Virginie Hamel & Paul Guichard Lab at University of Geneva
#cryoEM/ET❄️ and #UExM ⚗️ #ExpansionMicroscopy #TeamTomo
Genève, Suisse 🇨🇭
https://mocel.unige.ch/research-groups/guichard-hamel/overv
Y. Eva Tan Professor in Neurotechnology, MIT. Investigator, HHMI. Leader, Synthetic Neurobiology Group, http://synthneuro.org. Scientist, inventor, entrepreneur.
Postdoc in @cjrosenlab.bsky.social at MaineHealth 🦴🔬
Postdoc in Orion Weiner's lab at UCSF. I work at the interface of Cell Biology and Biophysics.
I am into: #Actin, #Membrane, #CellMigration, #Polarity, #Cats, #Mechanobiology #Cheese #CellBiology
Scientist and Global Sales Director at 3i (@the.3i.social), foodie, family man. Tai Chi and meditation matter. Compassion Heals. Microscopes Rock. https://www.intelligent-imaging.com
Studying the cell nucleus in search of inspiration for future DNA computers. Systems Biology professor at Karlsruhe Institute of Technology, post opinions mine alone.
hilbertlab.org
physics of life, biophysics, systems biology researcher at the University of Edinburgh
Actin, imaging, neurodegenerative disease. World record holder for Phish shows attended by a cell biologist. Associate Professor at MCG-Augusta University.