
Help You Help Future You: Organizing your research projects reproducibly
It's that time of year again – it's time to start a new research project! Planning for the research itself can be daunting, but planning for your work to be fully reproducible is a whole other layer t...
📣 New blog post!
Starting a new scientific research project? Make life easier for your future self! Check out this guide from Data Lab on organizing your code, files, and workflows so your research stays reproducible.
#ReproducibleResearch #OpenScience
03.12.2025 15:19 — 👍 0 🔁 0 💬 0 📌 0
📣 Applications are open for our final workshop of the year!
If you're a #PediatricCancer researcher looking to build on your foundational skills in #SingleCell #RNASeq analysis, this workshop may be for you.
🎓 Topic: Advanced Single-cell RNA-Seq
📅 Date/Time: December 8-12, 2025
📍 Virtual
09.10.2025 14:56 — 👍 0 🔁 0 💬 1 📌 0

An example of downloading a project from the ScPCA Portal.
📊 Access rich datasets that include data, metadata, and expertly curated cell type annotations…for free!
30.09.2025 14:00 — 👍 0 🔁 0 💬 0 📌 0
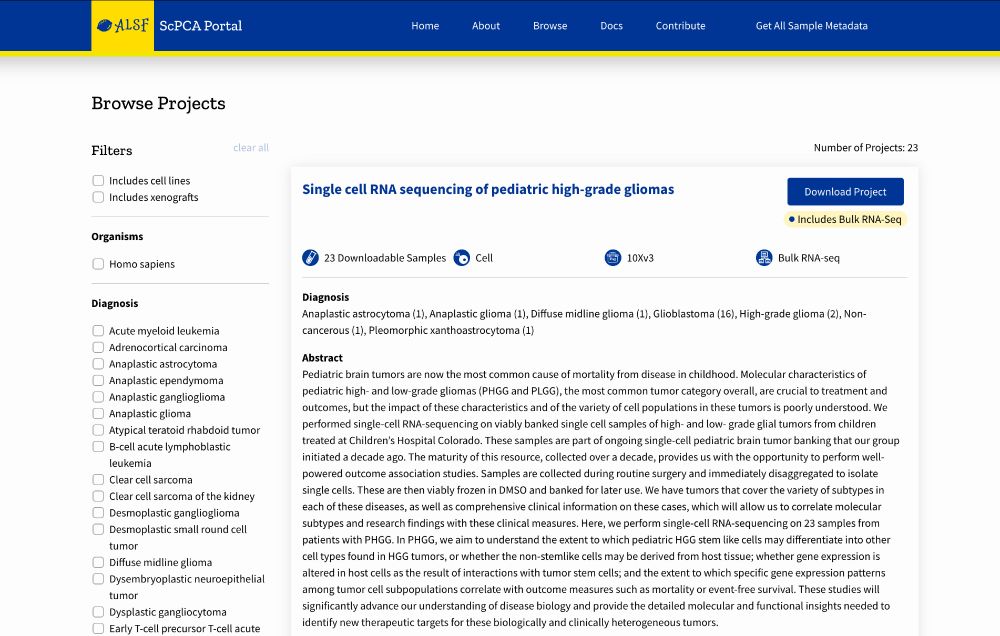
A project page on the ScPCA Portal.
🔍 User-friendly search and filter tools allow you to quickly find the projects you’re interested in.
30.09.2025 14:00 — 👍 0 🔁 0 💬 1 📌 0

Homepage of the Single-cell Pediatric Cancer Atlas (ScPCA) Portal.
🔬Explore over 700 samples from 55 pediatric cancer types, all in one place.
30.09.2025 14:00 — 👍 0 🔁 0 💬 1 📌 0
Attending the NCI Office of Data Sharing Annual Symposium? Stop by our table during poster/demo sessions for a live look at the Single-cell Pediatric Cancer Atlas Portal (scpca.alexslemonade.org).
📆 September 30 (today!) & October 1
📍 Poster/demo sessions
#SingleCell #PediatricCancer #DataSharing
30.09.2025 14:00 — 👍 0 🔁 0 💬 1 📌 0

Connect with our team at the #AACR Special Conference: Discovery and Innovation in #PediatricCancer! Dr. Ally Hawkins will be presenting our work on a collaboration aimed at uncovering therapeutic vulnerabilities in #EwingSarcoma.
🗓️ Tonight (9/25) | 7-9:30PM
📍 Poster Session A
25.09.2025 16:41 — 👍 1 🔁 0 💬 0 📌 0
🗓️ September 25 | 7-9:30 PM
📍 Poster Session A
🧾 "ews-nf: A custom workflow for tumor cell annotation and analysis of single-cell RNA-sequencing of paired patient Ewing sarcoma specimens from the Sean Karl cohort"
Connect with us at AACR and explore how we can support your research too!
23.09.2025 16:10 — 👍 2 🔁 0 💬 0 📌 0
Are you attending the #AACR Special Conference: Discovery and Innovation in #PediatricCancer? Stop by our poster to learn how we're collaborating with other labs to tackle their complex data challenges! 🧵
23.09.2025 16:10 — 👍 1 🔁 0 💬 1 📌 0

Pediatric Cancer Researchers Driving Progress Through Data, Training, and Collaboration
At the Childhood Cancer Data Lab, we support pediatric cancer researchers by providing data science training, collaborating on data-intensive projects, and designing open-source tools. Through this wo...
September is #ChildhoodCancerAwarenessMonth, a time to recognize the urgent need for better treatments and outcomes for kids with cancer. In our latest blog, we’re highlighting a few researchers who have drawn on the Data Lab’s resources in their remarkable efforts to accelerate new cure discovery!
19.09.2025 15:30 — 👍 0 🔁 1 💬 0 📌 0
Are you presenting at any of these events? Drop your poster or demo info in the comments. We hope to see you there!
08.09.2025 15:52 — 👍 0 🔁 0 💬 0 📌 0
📍 NCI Office of Data Sharing Annual Symposium
Learn how to access and use pediatric cancer data that may align with your research interests! Catch our live demo of the #SingleCell #PediatricCancer Atlas (ScPCA) Portal.
📆 Stop by our table during poster/demo sessions on September 30 and October 1!
08.09.2025 15:52 — 👍 0 🔁 0 💬 1 📌 0
📍 #AACR Special Conference in Cancer Research - Discovery and Innovation in #PediatricCancer
We’re showcasing an ongoing collaboration, supporting a multi-institutional effort to understand the therapeutic vulnerabilities of #EwingSarcoma.
📆 Poster Session A on September 25 (7-9:30 PM).
08.09.2025 15:52 — 👍 0 🔁 0 💬 1 📌 0
This fall, the Data Lab is hitting the road to present some of our tools and projects during these exciting events. Here’s where you can find us! 🧵
08.09.2025 15:52 — 👍 0 🔁 1 💬 1 📌 0
What can #PediatricCancer researchers expect from a Data Lab workshop?
✅ Interactive, hands-on training
✅ Instructors who take the time to understand your research goals
✅ Ongoing support with opportunities for 1:1 consultation, even after the workshop ends!
20.08.2025 14:55 — 👍 0 🔁 0 💬 1 📌 0

Level Up Your Reproducible Research Skills at Our Next Workshop!
At the Childhood Cancer Data Lab, we’re committed to helping pediatric cancer researchers work more efficiently, collaboratively, and reproducibly. That’s why we created our Reproducible Research Prac...
Whether you're looking to set up efficient processes in your lab, improve your ability to share data and code, or manage a collaborative project, this workshop can set you up for success.
🚀 Read our blog for full details and to apply!
04.08.2025 14:01 — 👍 0 🔁 0 💬 0 📌 0
Our #ReproducibleResearch Practices workshop teaches tools and techniques to help #PediatricCancer researchers work more efficiently, collaboratively, and reproducibly. The next course is taking place November 6–7, 2025, in Philadelphia, PA. 🧵
04.08.2025 14:01 — 👍 0 🔁 0 💬 1 📌 0
🚀 Easily download 700 samples from over 55 pediatric cancer types at scpca.alexslemonade.org!
24.07.2025 16:26 — 👍 0 🔁 0 💬 0 📌 0
Here’s what you’ll get with each dataset you download from the #SingleCell #PediatricCancer Atlas (ScPCA) Portal…for free!
-Unfiltered, filtered & processed counts
-QC report
-Associated metadata
-Reliable cell type annotations
24.07.2025 16:26 — 👍 0 🔁 0 💬 1 📌 0
📣 Join the Open #SingleCell #PediatricCancer Atlas (OpenScPCA) project. Grants are available!
We aim to annotate cell types for 19 projects on the ScPCA Portal. Explore the grant table to see which cancer types need your insight: github.com/AlexsLemonade/OpenScPCA-analysis/discussions/1062.
16.07.2025 15:48 — 👍 0 🔁 0 💬 0 📌 0
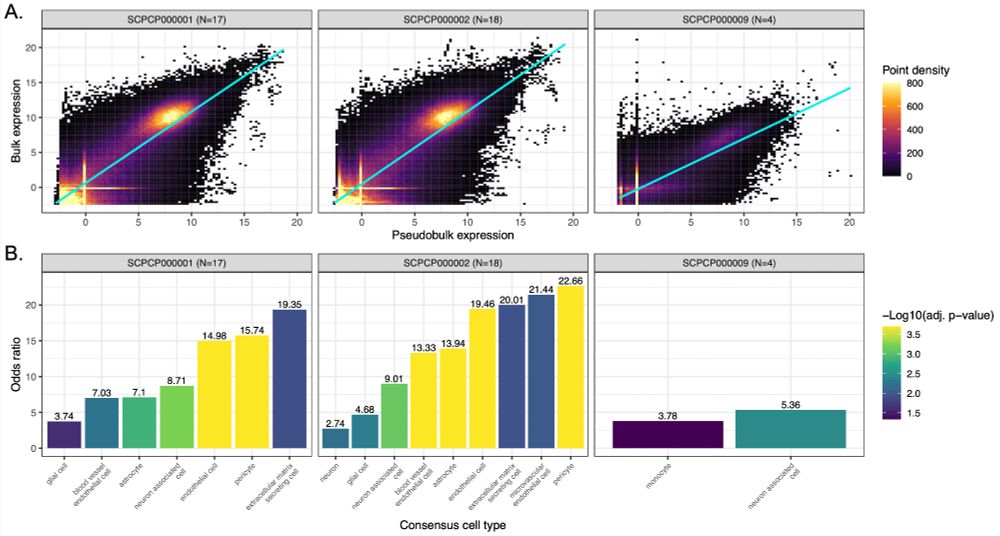
Figure 6 from the ScPCA preprint: Comparison of bulk and pseudo-bulk modalities.
Several projects on the Portal contain bulk RNA-seq data in addition to single-cell/nuclei RNA-seq data. We compared modalities across five solid tumor projects to explore potential biological differences.
01.07.2025 17:06 — 👍 0 🔁 0 💬 1 📌 0
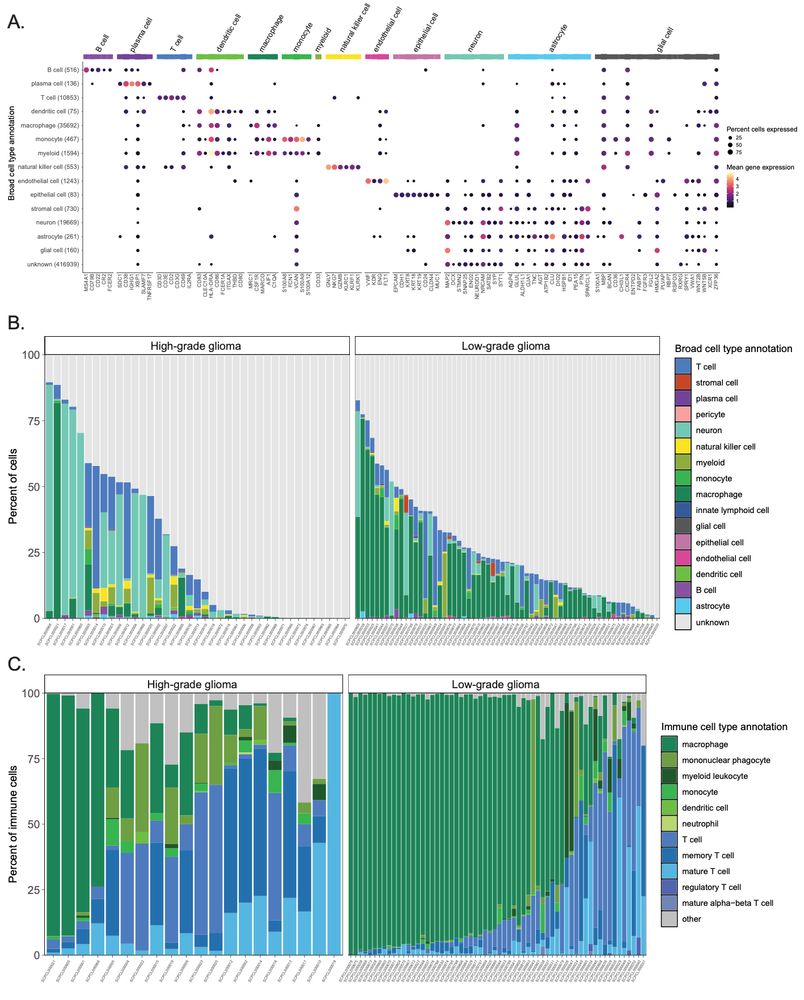
Figure 5 from the ScPCA preprint: Consensus cell type annotations in Brain and CNS tumors.
Using an ontology-based approach, we have assigned consensus cell types to all samples on the ScPCA Portal. They are now included in all processed and merged datasets from the Portal!
01.07.2025 17:06 — 👍 1 🔁 0 💬 1 📌 0
We’ve made exciting updates to the #SingleCell #PediatricCancer Atlas (ScPCA) Portal, making the available data even more robust, reliable, and useful for the research community! 🧵
01.07.2025 17:06 — 👍 0 🔁 1 💬 1 📌 0
Upcoming training opportunities!
-Reproducible Research Practices: November 2025
-Advanced Single-cell RNA-Sequencing: December 2025
Are you are a #PediatricCancer researcher interested in attending one of these workshops? Sign up to be notified when applications open!
✉️: bit.ly/3eQZXGQ
12.06.2025 14:30 — 👍 0 🔁 1 💬 0 📌 0

Thank you to everyone who joined us to learn advanced topics in #SingleCell #RNAseq analysis this week. We look forward to hosting more researchers at our workshops this year.
12.06.2025 14:30 — 👍 1 🔁 1 💬 1 📌 0

Building Reproducible Research Skills: A Training Workshop with the Treehouse Childhood Cancer Initiative
The Data Lab recently traveled to California to lead a hands-on workshop for nine researchers from the UC Santa Cruz Treehouse Childhood Cancer Initiative. The participants, all from a range of backgr...
We teamed up with the Treehouse Childhood Cancer Initiative at @ucscgenomics.bsky.social to deliver a custom workshop on reproducible research. In our new blog, Dr. Holly Beale, Lead Computational Biologist at Treehouse, shares how their team is putting new skills into practice. Read the full story!
04.06.2025 16:49 — 👍 4 🔁 1 💬 0 📌 0
Excited about science, immunology, tumour microenvironment and new therapies.🇫🇷🇬🇧🇪🇺 ❤️🔬
PhD Student @templemedschool | MS in Biotech @keanstem | BS in Exercise Sci @RutgersSAS | AS in Biology @raritanvalleycc | #microenvironment #cancer #lungcancer #phdlife
First Nations childhood cancer research | brain cancer | Kamilaroi | beach lover | toddler mum
Rotating Curator for the @rladies.org community💜
🔗 https://guide.rladies.org/rocur/
This week's curator: TBA
A journal dedicated to publishing the latest advances across all areas of cell biology. Part of @natureportfolio.nature.com
nature.com/ncb/index.html
Ph.D. Candidate at The Ohio State University and Nationwide Children’s Hospital
Empowering Computational Biology, Transforming Global Science!
The International Society for Computational Biology is a scholarly society for advancing understanding of living systems through computation, and communicating scientific advances worldwide.
The leading provider of open source workflow orchestration software for data pipelines, cloud infrastructure & collaboration. From the creators of @nextflow.io and @multiqc.info
Entertaining & educational conversations about science, tech, + more. Hosted by Ira Flatow and Flora Lichtman. From WNYCStudios.
Research scientist studying JMML @ UCSF
Science Writer. Childhood Cancer Advocate. Gardens. Global Conflict. JHU. Temple U. *All views expressed are my own*
Our mission: raise awareness and fund research to detect and treat cancers with the aspiration to cure all patients. linktr.ee/su2c
This account is to help populate the PedSky feed, but the feed is transtioning to the MedSky PedSky feed. Like and Pin https://bsky.app/profile/did:plc:fmug4zegnl7lf6ljp7aokpeh/feed/pediatrics
Research Software Engineer with the Department of Biomedical Informatics at the University of Colorado Anschutz Medical Campus
The Carpentries: Software Carpentry, Data Carpentry and Library Carpentry. An open global community teaching the skills and perspectives to turn data into knowledge
Pediatric oncologist. Cancer clinical epi and novice ImpSci researcher. Justice enthusiast. Academic freedom proponent. She/her.
Pediatric Psychologist at CHOP Cancer Center. Associate Professor at UPENN School of Medicine. Tar Heel bred. Beginning 🪕player. He/him
Founded in 1981, ASPHO is the only professional medical society dedicated solely to pediatric hematology/oncology subspecialists, supporting over 2,000 members. Learn more at aspho.org.
Focusing the power of genomics to create a healthier world. Leader in pangenomics, conservation genomics, cancer genomics, pathogen genomics, and nanopore sequencing. Home of the UCSC Genome Browser, @ucscxena, UShER, Dockstore, and other high-power tools.












