The paper’s great too 😅
07.02.2026 18:03 — 👍 0 🔁 0 💬 0 📌 0
I love that movie. There’s something very satisfying about watching people do crazy amazing things from the safety of your sofa
07.02.2026 18:03 — 👍 0 🔁 0 💬 1 📌 0
Woo!!! @hiwwright.bsky.social bamboozles us with his All of Us mastery!
This is the result of lot of hard work and some brilliance from one of the most talented ECRs I’ve worked with
A very important message which will reduce a lot of noise in aggregate associations, and improve interpretability
06.02.2026 10:01 — 👍 3 🔁 0 💬 0 📌 0
What genetic change to Indian wheat led to superior baking in a tandoor?
A naan sense mutation
10.01.2026 00:07 — 👍 170 🔁 25 💬 18 📌 6

Our new study defines a distinct #neurogenetic condition arising from recurrent structural variants at 16p13.3 palindrome.
Individuals show progressive ataxia, cognitive decline, and a characteristic MRI pattern with caudate & cerebellar atrophy.
#Genomics #RareDisease 🧵1/3
08.12.2025 08:41 — 👍 14 🔁 7 💬 2 📌 0
“Herasight claims to deliver an average gain of six IQ points for a couple with five embryos”
Probably with a sd of 20 😂
Paying £40k for that is insanity
06.12.2025 11:09 — 👍 1 🔁 0 💬 0 📌 0
Early Christmas present! Thank you Father Quistmas!🎅
02.12.2025 13:31 — 👍 3 🔁 0 💬 0 📌 0
Domain-wide Mapping of Peer-reviewed Literature for Genetic Developmental Disorders using Machine Learning and Gene2Phenotype https://www.medrxiv.org/content/10.1101/2025.11.24.25340871v1
25.11.2025 20:40 — 👍 3 🔁 2 💬 0 📌 0
It was a privilege to be part of this important international effort. How should we determine the value of genomics in healthcare?
27.11.2025 19:12 — 👍 6 🔁 2 💬 0 📌 0
Please forward this to any you know who could be interested!
The incredible @drghawkes.bsky.social will be leading a course on analysis of genome sequence data, functional annotation of the genome, and using the very, very exciting AlphaGenome tool
17.11.2025 14:02 — 👍 2 🔁 1 💬 0 📌 0
Great work Alex and team! Have been following with interest for a while.
I’m still in team “2nd tier test” 😅 but all of your work has made me think about it more.
Do you think you will make much gain using pangenome assembly, in particular for complex gene/regions?
17.11.2025 10:48 — 👍 0 🔁 0 💬 0 📌 0
Congrats @hilarycmartin.bsky.social!!
14.11.2025 20:40 — 👍 1 🔁 0 💬 0 📌 0
Thank you 🙂 I’m looking forward to reading your flexRV paper and giving it a try!
13.11.2025 18:39 — 👍 0 🔁 0 💬 0 📌 0
Rarely in the entire history of science, has QC been a topic of such passion, importance and impact. If only the French and Americans had adopted such rigor when they messed up the design of that multi billion $ telescope because one was using the metric system, the other the imperial system.
08.11.2025 15:35 — 👍 6 🔁 3 💬 0 📌 0
hell yes! (another quote for you kartik)
10.11.2025 09:15 — 👍 3 🔁 1 💬 1 📌 0
Genomic superstar @chundru.bsky.social taking on fake-news genotypes in >900k individuals. He shows allele-level filtering is rarely suffifient, and makes the brave choice to properly tackle chrX!
We’ll be providing our filtered AoU WGS plink pgens for all registered users: watch this space
10.11.2025 13:10 — 👍 7 🔁 4 💬 0 📌 0
Ha! Thanks, I'll file right under "Even reviewer 3 was speechless"
10.11.2025 11:57 — 👍 2 🔁 0 💬 0 📌 0
Thank you to other co-authors @carolinefwright.bsky.social, @mnweedon.bsky.social, @timfrayling.bsky.social, and @drarwood.bsky.social, @nihrexeterbrc.bsky.social, biobanks @ukbiobank.bsky.social and All of Us, and all of the participants of the studies
08.11.2025 09:31 — 👍 4 🔁 1 💬 1 📌 0
A massive, massive thank you to @hiwwright.bsky.social, @rnbeaumont.bsky.social, @drghawkes.bsky.social who all really drove this project to completion. Without them I would still be twiddling my thumbs shouting to the clouds about QC (I still will, but now you can read about it too!)
08.11.2025 09:31 — 👍 5 🔁 2 💬 1 📌 0

GitHub - chundruv/ukbb_pvcf2pgen: UKBB pVCF to plink pgen conversion
UKBB pVCF to plink pgen conversion. Contribute to chundruv/ukbb_pvcf2pgen development by creating an account on GitHub.
Using our DNANexus applet it is fast and not too expensive to QC and convert the entire UK Biobank WGS files to pgens! github.com/chundruv/ukb...
And for All of Us v8, we will provide you with the QC’ed pgen files on publication in a public workspace available to registered users.
08.11.2025 09:31 — 👍 4 🔁 1 💬 1 📌 0
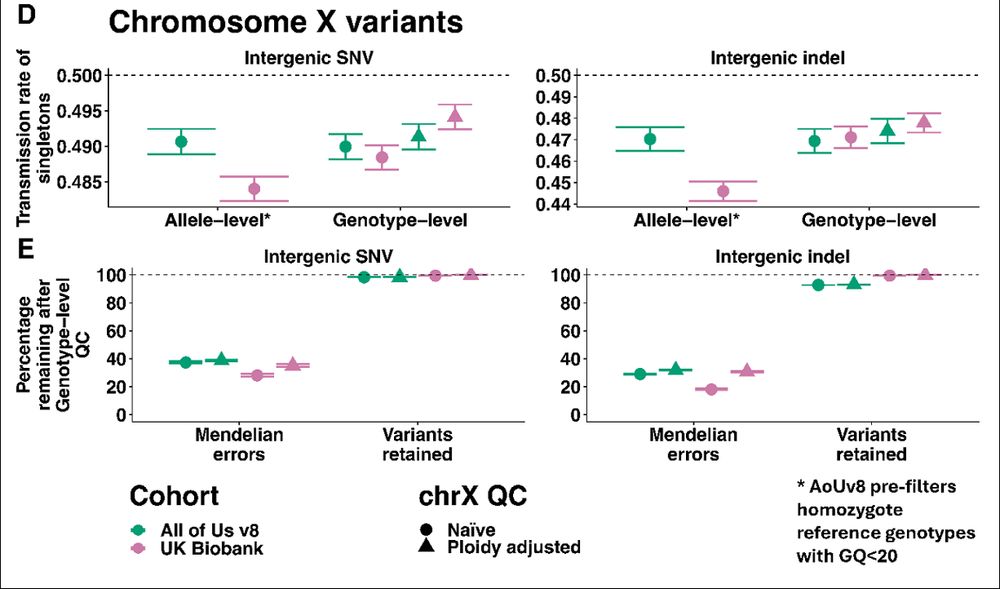
“Ok fine, but what about the X chromosome, you always forget that”
This time we didn’t ignore the X chromosome! We show that you should pay special attention to non-pseudoautosomal X chromosome where QC should be more lenient for haploid males.
08.11.2025 09:31 — 👍 3 🔁 1 💬 1 📌 0

“But Kartik, how do we know the genotypes are wrong?”
Trios! Both cohorts have ~1k parent-offspring trios that were recruited incidentally.
Applying genotype-level QC reduces Mendelian errors by ~60-80% (even in All of Us where they already did genotype-level QC on hom-refs!)
08.11.2025 09:31 — 👍 3 🔁 2 💬 1 📌 0

“Bah humbug! How bad could it be?”
After genotype-level QC and a 10% missingness cut-off, we remove ~100 million (~9%) variants!
Most genotypes removed are homozygote reference (which were filtered in All of Us already)
08.11.2025 09:31 — 👍 4 🔁 1 💬 1 📌 0

We caution that the released data in UK Biobank and All of Us is not as clean as you may believe!
Here, we show how we determine data quality in WGS data, provide a really fast way of doing so on biobank data, and we will release QC’ed plink files for All of Us upon publication
08.11.2025 09:31 — 👍 3 🔁 1 💬 1 📌 0
Really cool work from @jacquesml.bsky.social and @kash-a-patel.bsky.social 🥳
24% common variant heritability for a rare disease (MODY)!
This very elegantly shows the overlap between MODY and T2D, and hints at some potential MODY phenocopies
09.09.2025 18:50 — 👍 4 🔁 1 💬 0 📌 0
New preprint out now! We show polygenic background shapes GCK-MODY clinical presentation. In >1,000 cases, higher polygenic risk increased the chance of exceeding diagnostic diabetes thresholds, highlighting how monogenic & polygenic factors jointly shape disease. #Genetics
11.08.2025 16:57 — 👍 9 🔁 6 💬 1 📌 0

⌛Just over a week left!
Submit your abstract for the Cardiac Precision Medicine in the 21st Century Conference 🫀
📅 Oct 29–31, 2025 | Toronto
Showcase your research + compete for Best Abstract Award!
🔗 cardiacprecisionmedicine.com
#CardiacPrecision #Genomics #callforabstracts
27.06.2025 14:28 — 👍 2 🔁 2 💬 0 📌 0
PhD candidate//BCMM trainee ambassador
Lynch lab UCSF
studying the human oral microbiome in the context of periodontal disease and health
🪥🦠🫡🏳️🌈
Currently lurking.
Missed my dose of statistical genetics and cool methods.
AMJ: #1 open access destination for HCPs. Peer-reviewed journals & diverse content hub. Interviews, podcasts, webinars, infographics.
PhD-ing / @jmarshlab.bsky.social
Clinical Geneticist living in Edinburgh | Interested in Science, Genetics, Medicine
PhD Student at University of St Andrews
https://neurogenetics.st-andrews.ac.uk/
Postdoc at Globe Institute, University of Copenhagen 🇩🇰 she/her
Interested in the phenotypic effects of archaic DNA present in modern human genomes 🧬
Executive Editor/Team Lead Open Access Science & Medicine Journals Sage Publishing
Opinions = mine
http://linkedin.com/in/jlovick-editor
#oncology #cancerresearch #medicine #biology #cardiology #neurology #microbiology #publichealth #healthcare
Graduate Research Assistant at the University of Exeter
Current focus on cross-ancestry & partitioned SNP heritability:)
Clinical Genetics physician in West of Scotland and informatics/ML researcher at University of Edinburgh
Professor, Department of Medicine and McDonell Genome Institute @ Washington University. Specializing in Bioinformatics, Genomics, and Cancer. http://griffithlab.org/
Associate Professor at Erasmus MC. MD, PhD, Clinical Geneticist, interested in gene regulation and the non-coding genome, bridging research and patient care
Scientist at Illumina using AI methods to interpret the non-coding genome and empower new genetic association discoveries.