The lac operon in uropathogenic Escherichia coli enhances intracellular growth by enabling host glycan utilization https://www.biorxiv.org/content/10.64898/2025.12.03.691977v1
04.12.2025 03:16 — 👍 3 🔁 1 💬 0 📌 0
Overall, given that FOS are often included in infant formulas and used widely as prebiotics, it’s important to understand the mechanisms by which microbes utilize these oligosaccharides of different chain lengths
16.11.2025 01:48 — 👍 1 🔁 0 💬 0 📌 0
Very cool print demonstrating how variation in at TonB-dependent transporter repertoires shapes the ability of Bacteroides and Phocaeicola species to utilize fructooligosaccharides (FOS) of different chain lengths
16.11.2025 01:48 — 👍 12 🔁 7 💬 1 📌 0
Some cool data on bifidobacteria and their relatives in honeybees!
14.10.2025 21:48 — 👍 0 🔁 0 💬 0 📌 0
Genomic studies in bifidobacteria often focus only on CAZymes. IMHO, in Gram-positives, transporters are the true gatekeepers of glycan metabolism. We need to prioritize improving their functional annotations and include them in metabolic reconstructions (7/7)
30.09.2025 09:01 — 👍 1 🔁 0 💬 0 📌 0

We demonstrated that phylogenetically closely related strains can exhibit substantial differences in HMO utilization, which is driven by subtle variations in HMO transporter genes. Species/subspecies names alone don’t tell the full story; one needs to look at gene content in individual strains(6/7)
30.09.2025 09:01 — 👍 0 🔁 0 💬 1 📌 0

Among the new pathways we uncovered was a xyloglucan degradation pathway that was present in rare B. catenulatum subsp. kashiwanohense strains and conserved in B. dentium and B. tsurumiense (5/7)
30.09.2025 09:01 — 👍 0 🔁 0 💬 1 📌 0

We validated phenotypic predictions for 30 bifidobacterial strains, achieving 94% accuracy. For example, we confirmed the unique ability of the new B. longum clade to grow on starch and pullulan, and described an unconventional B. adolescentis strain that can use 2’-fucosyllactose (4/7)
30.09.2025 09:01 — 👍 0 🔁 0 💬 1 📌 0

Our analysis revealed notable inter- and intra-species variability. Among notable findings was a new Bifidobacterium longum clade harboring pathways for starch, pullulan, and difructose dianhydride metabolism but lacking pathways for LNB/GNB, N-glycan, and human milk oligosaccharide utilization(3/7)
30.09.2025 09:01 — 👍 0 🔁 0 💬 1 📌 0

We reconstructed 68 glycan utilization pathways encoded in 3,083 bif genomes by looking at the distribution of 589 curated metabolic functions (transporters, CAZymes, etc). Several years of manual curation greatly improved the quality of functional gene annotations (>90% for transporters!) (2/7)
30.09.2025 09:01 — 👍 0 🔁 0 💬 1 📌 0
It’s been a bit over 2 months since the main results of my PhD work on reconstructing carbohydrate utilization pathways in human bifidobacteria were finally published; so I guess it’s a good time to update the previous thread (1/7) www.nature.com/articles/s41...
30.09.2025 09:01 — 👍 3 🔁 1 💬 1 📌 0

Among the new pathways we uncovered was a xyloglucan degradation pathway that was present in rare B. catenulatum subsp. kashiwanohense strains and conserved in B. dentium and B. tsurumiense (5/7)
30.09.2025 08:49 — 👍 0 🔁 0 💬 0 📌 0

We validated phenotypic predictions for 30 bifidobacterial strains, achieving 94% accuracy. For example, we confirmed the unique ability of the new B. longum clade to grow on starch and pullulan, and described a B. adolescentis strain that can use 2’-fucosyllactose (4/7)
30.09.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0

Our analysis revealed notable inter- and intra-species variability. Among notable findings was a new Bifidobacterium longum clade harboring pathways for starch, pullulan, and difructose dianhydride metabolism but lacking pathways for LNB/GNB, N-glycan, and human milk oligosaccharide utilization(3/7)
30.09.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
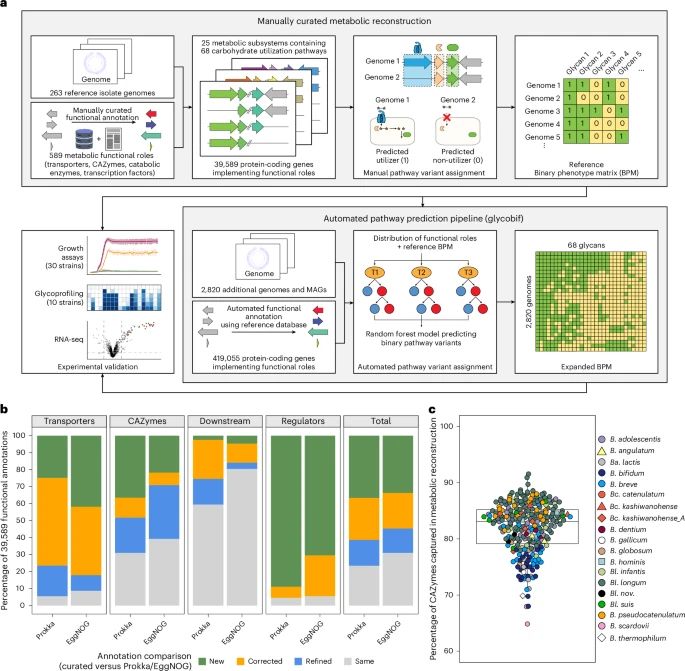
We reconstructed 68 glycan utilization pathways encoded in 3,083 bif genomes by looking at the distribution of 589 curated metabolic functions (transporters, CAZymes, etc). Several years of manual curation greatly improved the quality of functional gene annotations (>90% for transporters!) (2/7)
30.09.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
Excited to share our @cp-cellhostmicrobe.bsky.social led by Magda showing how Bif has co-evolved with different animal hosts 🐒🐭🐷🐦
Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
11.09.2025 14:49 — 👍 51 🔁 24 💬 0 📌 2
Our study developing a skin metatranscriptomics protocol is now out in @natbiotech.nature.com!
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
30.08.2025 02:17 — 👍 13 🔁 6 💬 1 📌 0
High-throughput single-cell isolation of Bifidobacterium strains from the gut microbiome https://www.biorxiv.org/content/10.1101/2025.07.23.666462v1
25.07.2025 04:17 — 👍 4 🔁 3 💬 0 📌 0

Identification and characterization of a sulfoglycosidase from Bifidobacterium bifidum implicated in mucin glycan utilization
Abstract. Human gut symbiont bifidobacteria possess carbohydrate-degrading enzymes that act on the O-linked glycans of intestinal mucins to utilize those c
Hi, very cool to see this out! I have a couple of comments about the model. 1) I think the GH20 enzyme that removes GlcNAc-6S (BbhII) is extracellular. The released GlcNAc-6S can be cross-fed to Bifidobacterium breve (some strains have a dedicated utilization pathway)
doi.org/10.1080/0916...
18.07.2025 16:46 — 👍 0 🔁 0 💬 1 📌 0
Postdoc @BorkLab.bsky.social @embl.org. PhD @UniHohenheim.bsky.social. Microbiome, HGT, FMT, Strain-level Metagenomics
Postdoctoral fellow, Massachusetts Institute of Technology
https://sites.google.com/view/smorenogamez
Associate Professor at Univ. of Naples Federico II, Italy - Machine learning, metagenomics, microbiome
Microbial Ecologist, Marine Biologist, PhD. She/Her. Views are my own.
https://cpavloud.github.io/mysite/
Researcher at Dept. CIBIO (Univ. of Trento, Italy), previously postdoc @cibiocm.bsky.social, love music and play keyboards.
Postdoc at Centre for Microbiome Research at QUT. Bioinformatics, metagenomics, Bin Chicken, permafrost...
🔬 Bioinformatician postdoc @oistedu.bsky.social working on microbiome & lab. automation.
🧪🦠 Proud member of @microbiomevif.bsky.social
scientist, traveller, climber, into drawing
Postdoc at Princeton 🐅
Trustee for AMI (http://appliedmicrobiology.org)
🌿ecology | 🧫 microbiomes | 🧬 genomics |
🦠 pathogen emergence | 💊 AMR | 👣 human impacts | 🐾 wildlife conservation |🌎 OneHealth
UNL & Stanford alum 🎓
💻 http://kate-lagerstrom.github.io
PhD, Computational Evolutionary Biologist working on AI, viral metagenomics and phylogenomics.
• Postdoc @ucalgary.bsky.social
• Previously @cibiocm.bsky.social
--| https://andrea-silverj.github.io/ |--
𝕏 : @silverjand
Microbiologist at UNIL Switzerland, interested in Microbiota-host interactions, symbiosis, evolution and genomics, and social bees
engelbeelab.com
I study evolution in fungi using genomics at UC Riverside. He/him.
Tongva, Cahuilla, Serrano lands
http://lab.stajich.org/
Bloomberg Distinguished Professor of BME, CS, and Biostats at Johns Hopkins Univ., tennis player, @StevenSalzberg1 on Twitter, lab: salzberg-lab.org, Substack blog: stevensalzberg.substack.com
A microbiologist 🦠& bioinformatician 🧬 uncovering diverse microbial mechanisms via AI ✨. A rising assistant professor @NTU Singapore. With data in the cloud, the sky is the limit! Check out our lab website :) https://genomiverse.net/
Microbiome researcher, co-leader of a family of soon five, Developer of metagenome-atlas.
silask.github.io
NIH Fogarty Fellow in Biomedicine @AgaKhanUniversity |Gates Funded Fellow in Microbiome @UniofBirmingham
Mentor, scientist & engineer. Having fun in @slavovlab.bsky.social and Parallel Squared Technology Institute @parallelsq.bsky.social with biology & single-cell proteomics.
https://nikolai.slavovlab.net
Computational microbiologist. Senior scientist at @cemess.bsky.social, @univie.ac.at.
Microbial ecology, mostly of nitrogen cycle microbes, and data driven physiology.
Maintainer of the GlobDB genome database https://globdb.org



















