
#ResearchExplained!
@shaonchakrabarti.bsky.social's lab at NCBS explored a new strategy to detect the internal clock’s control over the cell cycle, offering valuable insights that could enhance the effectiveness of chronotherapy-based cancer treatments.
Read Here: bit.ly/42X82Ax
✏️ Christeen Paulson
14.05.2025 10:11 — 👍 6 🔁 2 💬 0 📌 0
Our method opens up the possibility of discovering drug-tolerance related genes from single clinical samples, not just limited to settings where genetic engineering is feasible. Incredible work by #SuvranilGhosh, and a really fun and enjoyable collaboration with #ArchishmanRaju.
20.01.2025 10:50 — 👍 1 🔁 0 💬 0 📌 0
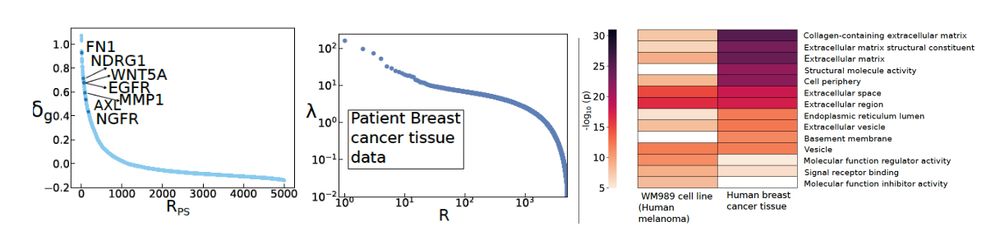
We demonstrated that memory genes discovered from many independent samples of a melanoma cell line (Memory-Seq), are recovered using Power-Seek, but with just one scRNA-seq dataset. Excitingly, we also demonstrate its applicability in human breast cancer tissue. Many ECM and EMT genes showed up.
20.01.2025 10:50 — 👍 1 🔁 0 💬 1 📌 0
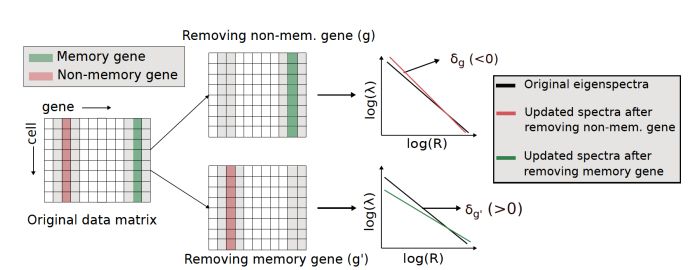
We showed that single samples will also exhibit power laws in the presence of memory genes. We developed a simple and easy to use algorithm, 'Power-Seek' (all puns intended). This detects memory genes using variations in power-law upon removing genes one at a time from a single scRNA-seq dataset.
20.01.2025 10:50 — 👍 0 🔁 0 💬 1 📌 0

Power law tails in phylogenetic systems | PNAS
Covariance analysis of protein sequence alignments uses coevolving pairs of sequence
positions to predict features of protein structure and functio...
Demonstrating the theory in single samples posed intriguing difficulties, since cell-cell correlations get mixed up with gene-level variations, precluding use of methods like PCA. This took us into a deep dive in Random Matrix Theory, from a beautiful paper on protein evolution: tinyurl.com/3y42v7zt
20.01.2025 10:50 — 👍 0 🔁 0 💬 1 📌 0
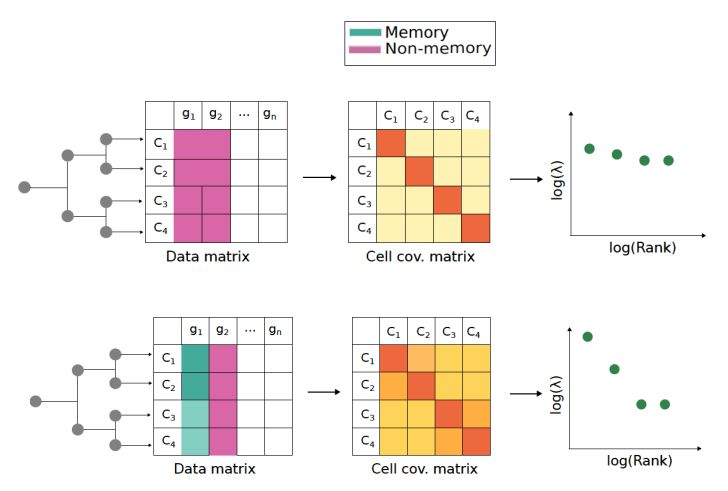
While seemingly impossible, this can indeed be done due to a beautiful underlying theoretical result that we show: memory genes, which give rise to lineage correlations, generate detectable Power-Laws in the eigenspectrum of the cell-covariance matrix of a scRNA-seq dataset.
20.01.2025 10:50 — 👍 0 🔁 0 💬 1 📌 0
However, these methods are conceptually based on variants of the classic Luria-Delbruck framework, requiring either many samples or lineage information from barcodes to identify memory genes. We asked whether *all* these requirements can be done away with using a completely different approach?
20.01.2025 10:50 — 👍 0 🔁 0 💬 1 📌 0
Identifying memory genes in cancer drug tolerance
Nature Reviews Cancer - In this Journal Club, Chakrabarti discusses a method to dissect the molecular architecture of inheritable gene expression (memory) states that mark cells that transition...
We now know that non-genetic states inherited across cell divisions are associated with cancer drug tolerance. Beautiful methods: Memory-Seq, Rewind, PATH, GEMLI have recently been developed to discover these states. We reviewed one such method (Rewind) recently: rdcu.be/d11Jm
20.01.2025 10:50 — 👍 0 🔁 0 💬 1 📌 0

Covering theory and experiments in chronobiology (including hands-on wet lab sessions), ranging from historical perspectives to more modern aspects of high-dimensional data analysis of bio-clocks, the workshop promises to be very exciting. Do apply by 31st December!
21.12.2024 15:44 — 👍 2 🔁 0 💬 0 📌 0
Identifying memory genes in cancer drug tolerance
Nature Reviews Cancer - In this Journal Club, Chakrabarti discusses a method to dissect the molecular architecture of inheritable gene expression (memory) states that mark cells that transition...
Our article on a cool technique to identify 'Memory Genes' associated with #Cancer #DrugTolerance is now out in
@NatureRevCancer! Also a sneak peek into work about to come out (collab with Archishman Raju) we are really excited about! rdcu.be/d11Jm
09.12.2024 14:57 — 👍 1 🔁 0 💬 0 📌 0
Cell biologist. Group leader/Wellcome CDA Fellow, Randall Centre at King’s College London
Assitant Professor ncbsbangalore.bsky.social Making dye based molecular tools for imaging, cell biology and proteomics. Previously @hhmijanelia.bsky.social @stonybrooku.bsky.social, IISER-Kolkata, and JNCASR. Lab news: DyeCraftLab.com
Heart of a developmental biologist, head of a mathematician/physicist. Posts from Tom and lab members.
https://twhiscock.github.io/
TIFR is a research institute & Deemed University for Natural Sciences, Mathematics, Computer Science & Science Education, under the Dept of Atomic Energy, Govt of India.
Website https://main.tifr.res.in/ | Science Communication https://tifr.res.in/scicomm
CRUK Career Development Fellow
Group Leader of @ccg-ucl.bsky.social at the UCL Cancer Institute, @CRUKLungCentre, and Visiting Scientist at the Francis Crick
W: http://www.ucl.ac.uk/cancer/zaccaria-lab
W: https://sites.google.com/view/ccgresearchgroup
Cancer Evolution and Genome Instability Lab Francis Crick Institute. Views my own
Exploring how our bodies tell time | McDonnell Fellow | Humboldt Fellow | INSA Young Scientist Medal | Herzog Lab | JNCASR 🇮🇳 - Charité 🇩🇪 - WashU 🇺🇸 | klnikhil.wordpress.com
The Anja Groth group in Copenhagen studies chromatin replication, epigenetic memory and (epi)genome stability in the context of mitotic cell division
https://vaikuntanathan-group.uchicago.edu
How do cells form tissues? Our research program spans the scales from molecules to organisms. News from the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) in Dresden, Germany. Impressum: https://www.mpi-cbg.de/impressum
In search of a platform that widens and deepens our understanding of the world. Less re-posting, more reading. Listening more, talking less. My opinions are my own. Some of them aren’t even mine.
Mom, Neuroscientist, Serotonin enthusiast, Professor at TIFR
Ecology and Evolution | Microbial Communities |Theory | Biophysics, Faculty member at ICTS
Theoretical Neuroscience, Deep Learning, & the space between.
Assistant Professor, Birla Institute of Technology & Science.
http://brain.bits-hyderabad.ac.in/venkat/
Science writer and author of books including Bright Earth, The Music Instinct, Beyond Weird, How Life Works.
Illuminating math and science. Supported by the Simons Foundation. 2022 Pulitzer Prize in Explanatory Reporting. www.quantamagazine.org
Associate Professor of Bioengineering, University of Washington. Interested in cell fate decisions, circuits and the immune system. The views expressed do not reflect those of my employer.
lab: depts.washington.edu/kuehlab/
Physics | Molecular Biology | Evolution.
Lecturer at UCL Physics.
Formerly:
Postdoc with @amurugan.bsky.social at the University of Chicago.
PhD with Madan Rao and Jitu Mayor at NCBS, Bangalore.
Advancing the frontiers of basic science through grantmaking, research and public engagement. Sign up for our newsletter: simonsfoundation.org/newsletter






