
You can read more about Anja's award here: novonordiskfonden.dk/en/news/biom...
14.01.2026 20:42 — 👍 10 🔁 3 💬 0 📌 1@grothlab.bsky.social
The Anja Groth group in Copenhagen studies chromatin replication, epigenetic memory and (epi)genome stability in the context of mitotic cell division

You can read more about Anja's award here: novonordiskfonden.dk/en/news/biom...
14.01.2026 20:42 — 👍 10 🔁 3 💬 0 📌 1
A picture of current members of the Groth lab
Congratulations to @groth-anja.bsky.social on receiving the Novo Nordisk Foundation Jacobæus Prize 🏆 we’re proud to see Anja's mentorship, leadership, & pioneering work in epigenetic cell memory getting top recognition 👏 celebrating the contributions of past & present lab members to this work too 🥳
14.01.2026 20:42 — 👍 67 🔁 11 💬 1 📌 0Our latest collaboration with the talented Susi @sbantele.bsky.social and Jiri Lukas is out now @science.org 🤩. We reveal the hidden price for DNA repair, with potential implications for genome function, gene therapy and ageing👇
07.11.2025 08:34 — 👍 43 🔁 16 💬 0 📌 1It was wonderful to hear talks from EpiC members @grothlab.bsky.social @nilskrietenstein.bsky.social @nilssonlab.bsky.social & Niels Mailand, now we are settled in our new home @cancer.dk. We are grateful for @dg.dk for this exciting opportunity. Onwards and upwards with EpiC science! 🧬 #CPRLegacy
28.08.2025 17:35 — 👍 4 🔁 0 💬 0 📌 0
Image of Niels Mejlgaar (CEO of DNRF), Anja Groth (EpiC director) & Jesper Fisker (CEO of DCS).
We have lift off!! 🚀 Yesterday was the official opening of EpiC! The DNRF Center for Epigenetic Cell Memory at the Danish Cancer Institute!
Our EpiC team with director Anja Groth @groth-anja.bsky.social, were joined by Jesper Fisker, CEO @cancer.dk & Niels Mejlgaar, CEO @dg.dk to celebrate 🎉

Less than two weeks left to apply!
Postdoc positions in my lab to study
1. initiation of DNA replication.
2. chromatin replication/epigenetic inheritance.
Great for biochemists, biophysicists & structural biologists.
Deadline 3 August 2025.
crick.wd3.myworkdayjobs.com/External/job...

Congratulations to Ada 🥳🍾, who very successfully defended her master's thesis today! You were an excellent student and we were super happy to have you in the lab! Wishing you all the best for the future and enjoy your well-deserved break!
27.06.2025 20:10 — 👍 10 🔁 0 💬 0 📌 0Postdoc positions open in my lab (Francis Crick Institute) to study
1. initiation of DNA replication.
2. chromatin replication/epigenetic inheritance.
Great for biochemists, biophysicists and cryo-EM/cryo-tomography scientists.
Deadline 3 August 2025.
crick.wd3.myworkdayjobs.com/External/job...
🚨 Postdoc wanted (🇳🇱NL based) for our exciting collaboration with the lab of Alexander van Oudenaarden @hubrechtinstitute.bsky.social in Utrecht, with collaborative visits to our lab Copenhagen 🇩🇰. We seek a computational scientist to model epigenetic inheritance, see more below 👇!
20.05.2025 10:20 — 👍 12 🔁 13 💬 0 📌 0Welcome Marcel!!
01.05.2025 19:31 — 👍 1 🔁 0 💬 1 📌 0Thank you, Tae-Kyeong!
10.03.2025 07:22 — 👍 1 🔁 0 💬 0 📌 0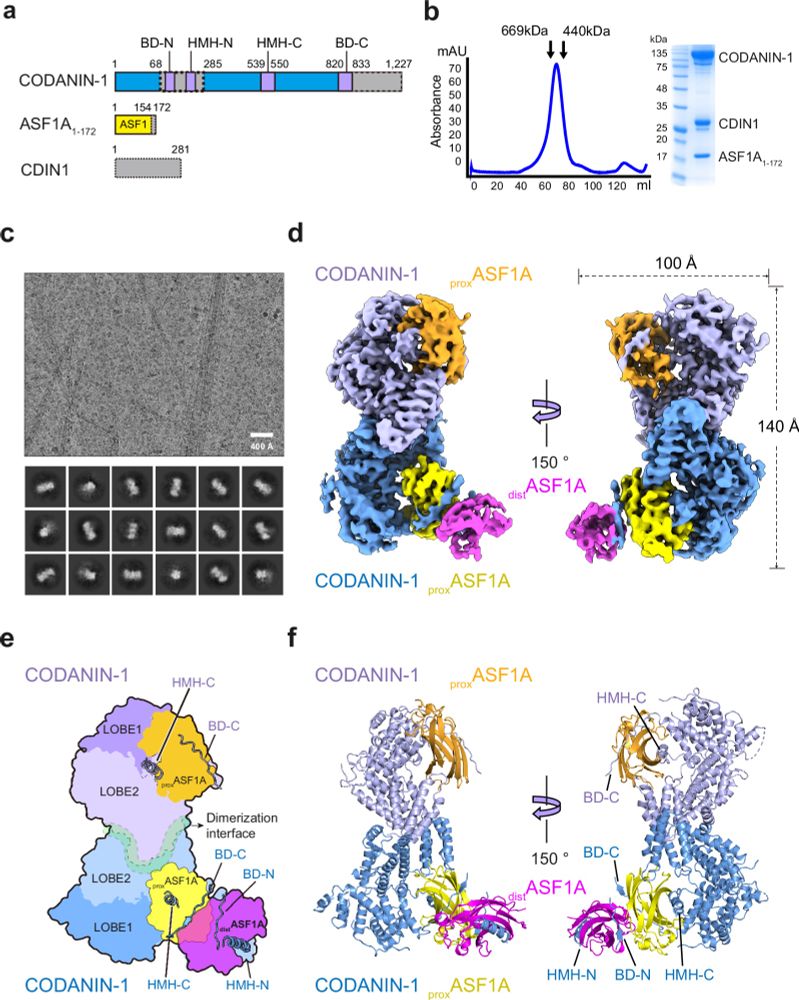
Together our work gives a molecular basis for histone supply regulation by CODANIN-1, read the full paper here: rdcu.be/ecbHq . Thanks to the Song lab at KAIST jijoonsong.bsky.social for taking us onboard as collaborators for this exciting new structure-function adventure!
06.03.2025 12:45 — 👍 1 🔁 0 💬 0 📌 0How important is CODANIN/ASF1A complex formation? Inhibiting dimerization in cells or introducing a pathogenic CDA-I mutation in the dimerization interface reduced ASF1A cytoplasmic sequestration. This shows the significance of CODANIN/ASF1A complex formation and possible disease relevance
06.03.2025 12:45 — 👍 0 🔁 0 💬 1 📌 0Which CODANIN binding sites are required for ASF1 regulation? Using inducible complementation of CODANIN-1 in cells, we found that mutation of both BD-N and HMH-C is needed to prevent cytoplasmic sequestration of ASF1A. Thus, CODANIN-1 regulates ASF1A nuclear import by this dual-binding mechanism.
06.03.2025 12:45 — 👍 0 🔁 0 💬 1 📌 0By binding in this way, CODANIN-1 occludes the two major histone-binding surfaces of ASF1A, preventing its chaperone activity. Interestingly, CODANIN-1 could also bind preformed ASF1A/H3-H4 using its B domains, suggesting that it may regulate ASF1 at multiple points in the histone supply chain.
06.03.2025 12:45 — 👍 0 🔁 0 💬 1 📌 0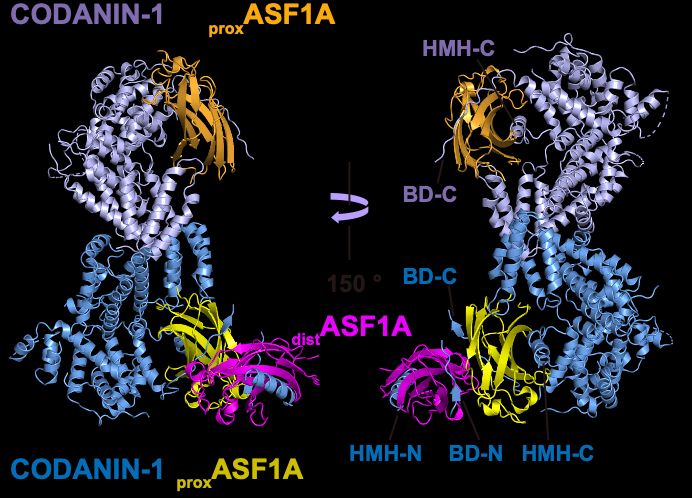
So, how does CODANIN-1 bind ASF1? Our cryo-EM structure reveals that CODANIN-1 forms a dimer, with each monomer binding two ASF1A molecules. These two ASF1As are held in proximal and distal positions by CODANIN’s N-terminal B domain (BD-N) and C-terminal histone H3 mimic helix (HMH-C), respectively.
06.03.2025 12:45 — 👍 0 🔁 0 💬 1 📌 0A bit of background: ASF1 plays a central role in histone nuclear import and supply for replication & transcription. CODANIN-1 is commonly mutated in a rare form of heritable anaemia (CDA-I) and inhibits histone supply by ASF1. But we don’t currently understand these roles on the molecular level.
06.03.2025 12:45 — 👍 1 🔁 0 💬 1 📌 0
𝐍𝐞𝐰 𝐦𝐞𝐜𝐡𝐚𝐧𝐢𝐬𝐭𝐢𝐜 𝐢𝐧𝐬𝐢𝐠𝐡𝐭𝐬 𝐢𝐧𝐭𝐨 𝐭𝐡𝐞 𝐫𝐞𝐠𝐮𝐥𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐡𝐢𝐬𝐭𝐨𝐧𝐞 𝐬𝐮𝐩𝐩𝐥𝐲! Excited to share our latest collaboration led by tkjeong.bsky.social and ciaranfrater.bsky.social! Structural determination of the histone chaperone ASF1 and its regulator CODANIN-1 uncovers how CODANIN-1 counters ASF1 activity. 🧵👇
06.03.2025 12:45 — 👍 55 🔁 14 💬 1 📌 2SAVE THE DATE! The next EMBO | EMBL Symposium on 'DNA replication and genome maintenance: from basic biology to disease' will be held October 20-23, 2026.
Organized by Helle Ulrich (IMB), Johannes Walter (Harvard) and Anja Groth (Danish Cancer Institute).
www.embl.org/about/info/c...
Epigenetics Gordon Research Conference. Consider joining us for what will be an excellent, exciting meeting and will be held in Europe for the first time: 10-15 Aug25 Barcelona, Spain. Please repost and apply early - this meeting is popular! www.grc.org/epigenetics-...
25.02.2025 16:54 — 👍 36 🔁 19 💬 1 📌 1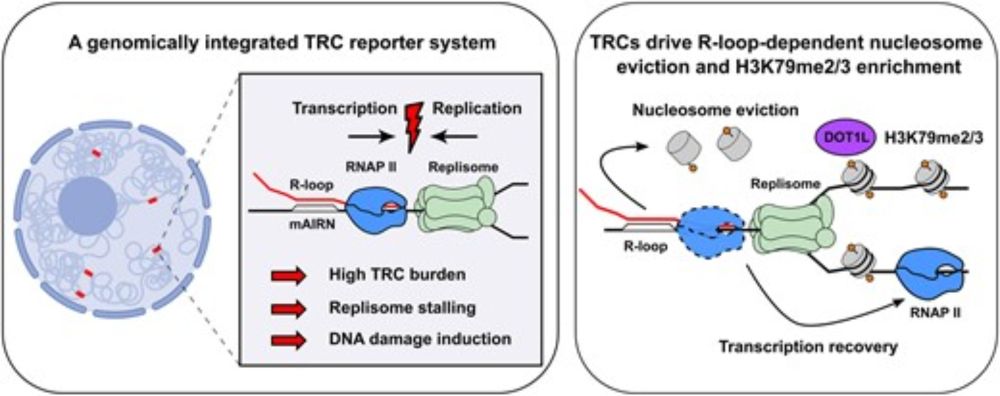
I am delighted to share the main work of my PhD project in the @hamperllab.bsky.social now out in @narjournal.bsky.social. TRCs drive R-loop-dependent nucleosome eviction and require DOT1L activity for transcription recovery.
academic.oup.com/nar/article/...
#TRC #r-loop #chromatin
More below 👇
Thank you, Jeroen!
20.02.2025 08:12 — 👍 0 🔁 0 💬 0 📌 0
If you’re interested in epigenetic inheritance and cell division, check out the vacancies for 6 PhDs and Postdocs positions in our new Center for Epigenetic Cell Memory (EpiC): tinyurl.com/yvd93na2 6/n
19.02.2025 19:35 — 👍 4 🔁 0 💬 0 📌 0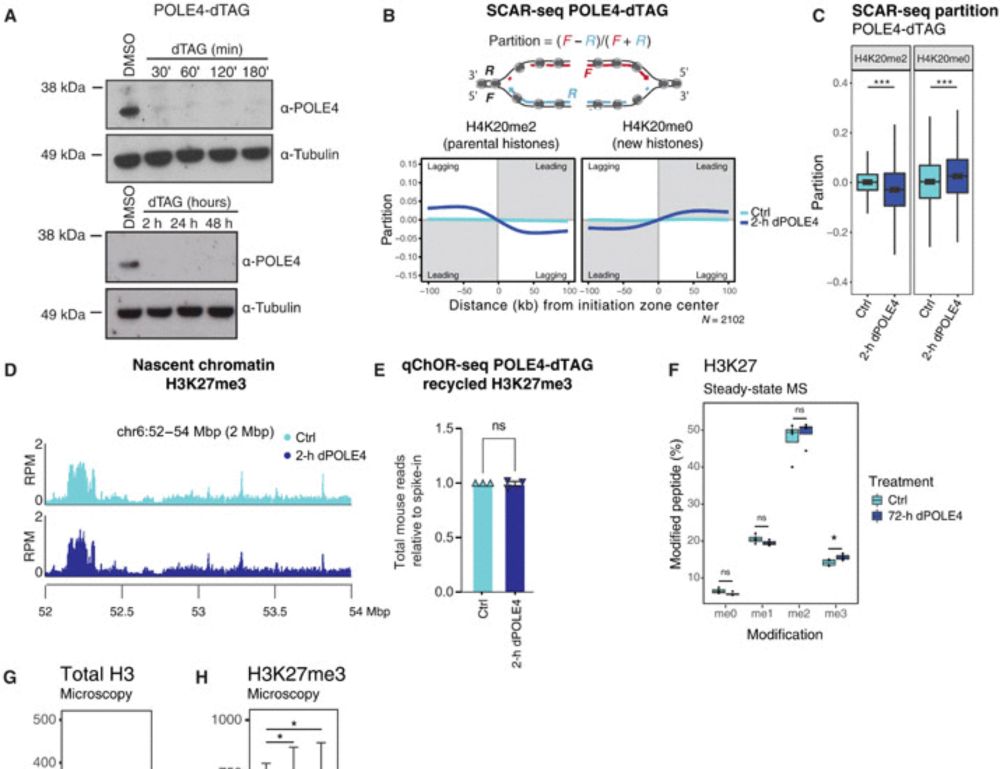
Want to dive deeper? 🧐 Read the full paper here: tinyurl.com/233y89d2 🔗📖. Last, a big thank you to all our collaborators and colleagues at #NNF-CPR 🙏. #HistoneRecycling #Epigenetics #Chromatin #ScienceAdvances 5/n
19.02.2025 19:35 — 👍 4 🔁 0 💬 2 📌 0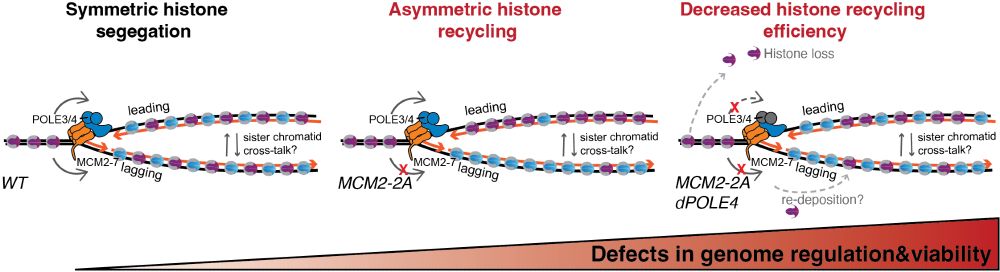
Our work highlights 𝐩𝐚𝐫𝐞𝐧𝐭𝐚𝐥 𝐡𝐢𝐬𝐭𝐨𝐧𝐞 𝐫𝐞𝐜𝐲𝐜𝐥𝐢𝐧𝐠 𝐚𝐬 𝐚 𝐤𝐞𝐲 𝐞𝐩𝐢𝐠𝐞𝐧𝐞𝐭𝐢𝐜 𝐬𝐚𝐟𝐞𝐠𝐮𝐚𝐫𝐝 – histone modifications must be 𝐞𝐟𝐟𝐢𝐜𝐢𝐞𝐧𝐭𝐥𝐲 transmitted to 𝐛𝐨𝐭𝐡 daughter strands for this to work! Could failures in this process contribute to developmental disorders, cancer, or aging? More to explore! 4/n
19.02.2025 19:35 — 👍 6 🔁 0 💬 1 📌 0What happens next? Gene misregulation & differentiation defects start to emerge. But when 𝐛𝐨𝐭𝐡 leading & lagging strand recycling are disrupted (POLE4 & MCM2-2A), 𝐬𝐨𝐦𝐞 𝐩𝐚𝐫𝐞𝐧𝐭𝐚𝐥 𝐡𝐢𝐬𝐭𝐨𝐧𝐞𝐬 𝐚𝐫𝐞 𝐥𝐨𝐬𝐭 𝐢𝐧 𝐭𝐫𝐚𝐧𝐬𝐦𝐢𝐬𝐬𝐢𝐨𝐧 —worsening the defects. Unlike single mutants, cells now start to die! 3/n
19.02.2025 19:35 — 👍 4 🔁 0 💬 1 📌 0We use a degron to deplete POLE4, a leading strand recycler, in WT or a background defective in lagging strand recycling (MCM2-2A). The first epigenome alteration upon acute loss of leading strand recycling is 𝐚𝐜𝐜𝐮𝐦𝐮𝐥𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐇𝟑𝐊𝟐𝟕𝐦𝐞𝟑 - 𝐰𝐢𝐭𝐡𝐢𝐧 𝟏-𝟐 𝐜𝐞𝐥𝐥 𝐜𝐲𝐜𝐥𝐞𝐬 - 𝐛𝐞𝐟𝐨𝐫𝐞 𝐭𝐫𝐚𝐧𝐬𝐜𝐫𝐢𝐩𝐭𝐢𝐨𝐧𝐚𝐥 𝐜𝐡𝐚𝐧𝐠𝐞𝐬! 2/n
19.02.2025 19:35 — 👍 4 🔁 0 💬 1 📌 0During replication, modified parental histones are transmitted to the daughter DNA strands - they are recycled. Recycled histones serve as agents of memory to maintain the epigenome in daughter cells! What happens when recycling is acutely disrupted? 1/n
19.02.2025 19:35 — 👍 4 🔁 0 💬 1 📌 0
𝐇𝐨𝐰 𝐝𝐨 𝐜𝐞𝐥𝐥𝐬 𝐫𝐞𝐦𝐞𝐦𝐛𝐞𝐫 𝐰𝐡𝐨 𝐭𝐡𝐞𝐲 𝐚𝐫𝐞 𝐚𝐟𝐭𝐞𝐫 𝐃𝐍𝐀 𝐫𝐞𝐩𝐥𝐢𝐜𝐚𝐭𝐢𝐨𝐧? Our new study “Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability” is out in 𝘚𝘤𝘪𝘦𝘯𝘤𝘦 𝘈𝘥𝘷𝘢𝘯𝘤𝘦𝘴. Led by @lleonie.bsky.social @biranalva.bsky.social 🧵 More below👇
19.02.2025 19:35 — 👍 111 🔁 53 💬 5 📌 5
6 PhD and postdoc positions in the new Center for Epigenetic Cell Memory in Copenhagen, Denmark. Interested in epigenetics and chromatin b
biology? Consider applying here: tinyurl.com/yvd93na2.
Please share!!!