Btw, if you hate your PhD you can just leave. You can transfer (I did), you can drop out. More people probably should.
28.03.2025 01:51 — 👍 50 🔁 3 💬 4 📌 0Jeffrey Ouyang-Zhang
@zhang-ouyang.bsky.social
ML + Bio (prev CV) http://jozhang97.github.io
@zhang-ouyang.bsky.social
ML + Bio (prev CV) http://jozhang97.github.io
Btw, if you hate your PhD you can just leave. You can transfer (I did), you can drop out. More people probably should.
28.03.2025 01:51 — 👍 50 🔁 3 💬 4 📌 0First they came for Columbia...
10.03.2025 01:43 — 👍 15 🔁 3 💬 0 📌 0ooh also very curious 👀
27.11.2024 14:06 — 👍 1 🔁 0 💬 0 📌 0Could you add me to this list?
21.11.2024 15:51 — 👍 0 🔁 0 💬 0 📌 0Implementation is extremely simple. If you are using ESM2, you're just one line of code away from upgrading to ISM's enhanced capabilities. (7/7)
🔖 paper www.biorxiv.org/content/10.1...
💻 github: github.com/jozhang97/ISM
🤗 huggingface: huggingface.co/jozhang97/is...
To conclude, ISM takes sequence-only input but produces structurally-rich representations. After all, the amino acid sequence is the only genetic information necessary for protein folding. Our structural loss better enables transformers to learn sequence-structure mapping. (6/7)
13.11.2024 00:40 — 👍 1 🔁 0 💬 1 📌 0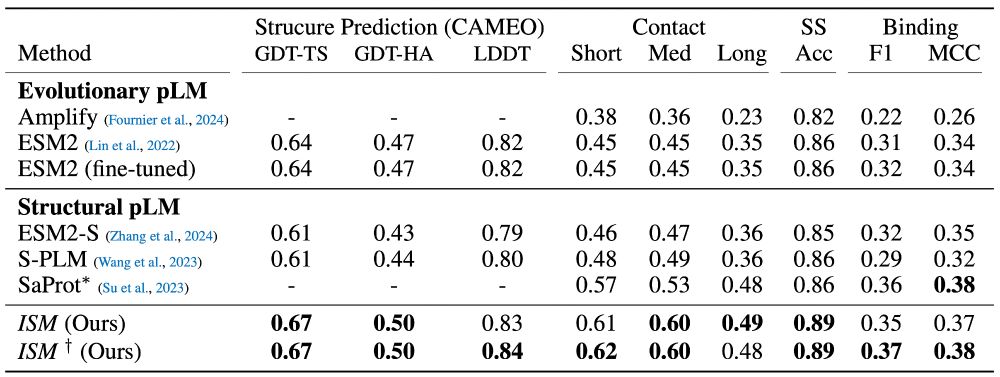
On structural benchmarks, we found that our model's structural representations outperform those from existing sequence models and even match performance with representations from models that take structure and sequence as input. (5/7)
13.11.2024 00:40 — 👍 0 🔁 0 💬 1 📌 0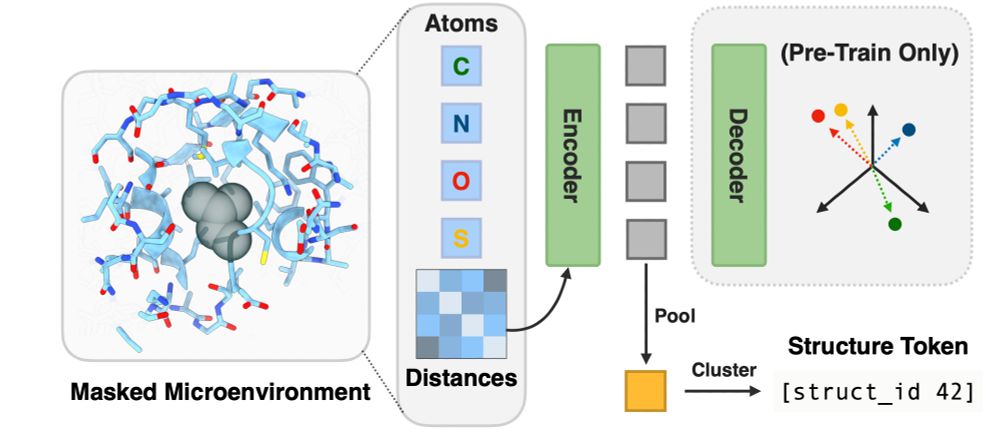
ISM's secret sauce is a microenvironment-based autoencoder. The all-atom autoencoder learns to embed the tertiary structure surrounding a residue into a structure token. We distill these per-residue tokens (and MutRank tokens) into ESM2. (4/7)
13.11.2024 00:40 — 👍 0 🔁 0 💬 1 📌 0Masked language modeling enables ESM2 to learn rich evolutionary features which capture a view of the structural landscape. However, it often underperforms structure models on downstream tasks.
We fine-tune ESM2 to predict representations from structure models. (3/7)
ISM is our latest protein language model which enhances ESM2 with enriched structural representations. (2/7)
13.11.2024 00:40 — 👍 0 🔁 0 💬 1 📌 0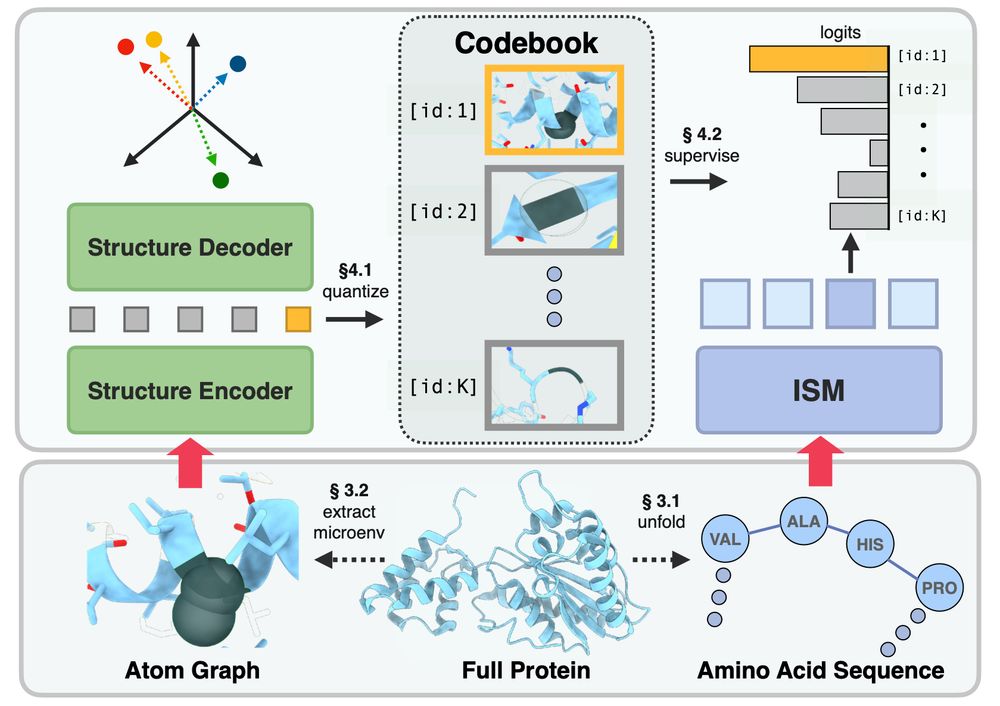
Are you using ESM2 for your sequence embeddings? Try out ISM, a one-line code change that will incorporate improved structure and sequence information, without a structure as input. (1/7)
13.11.2024 00:40 — 👍 5 🔁 0 💬 1 📌 0