
This week I presented DataSAIL at the #ISMBECCB2025 conference in #Liverpool
It has been an amazing chance and experience to meet many people working on information leakage. And getting great ideas to extend it
Nicely supervised by & colaborated with @dbblumenthal.bsky.social @ok55991.bsky.social
24.07.2025 16:03 — 👍 5 🔁 4 💬 0 📌 0
Very happy to see DataSAIL published in @naturecomms.bsky.social. Give it a try if you want to test if your ML models generalize to OOD scenarios. Great collaboration between @uni-saarland.de and @fau.de :-)
09.04.2025 08:27 — 👍 2 🔁 1 💬 0 📌 0

A graphic depicting people sitting at desks and working on computers in the foreground. In the background, there are more people standing around in an abstract space. The space itself looks futuristic.
Are you ready to challenge yourself and compete with the brightest minds in AI and computer science? 🧠 Join the FAU AI Innovation Challenge 2025! Compete for 10.000€ in prizes in various categories ranging from game AI, cybersecurity & more.
👉 Learn more: go.fau.de/1beg-
27.01.2025 12:34 — 👍 4 🔁 2 💬 0 📌 0
Deep learning models for sequence-based PPI prediction still fail to yield reliable predictions in challenging scenarios. Great work led by the amazing Timo Reim and @judith-bernett.bsky.social
27.01.2025 11:50 — 👍 3 🔁 0 💬 0 📌 0
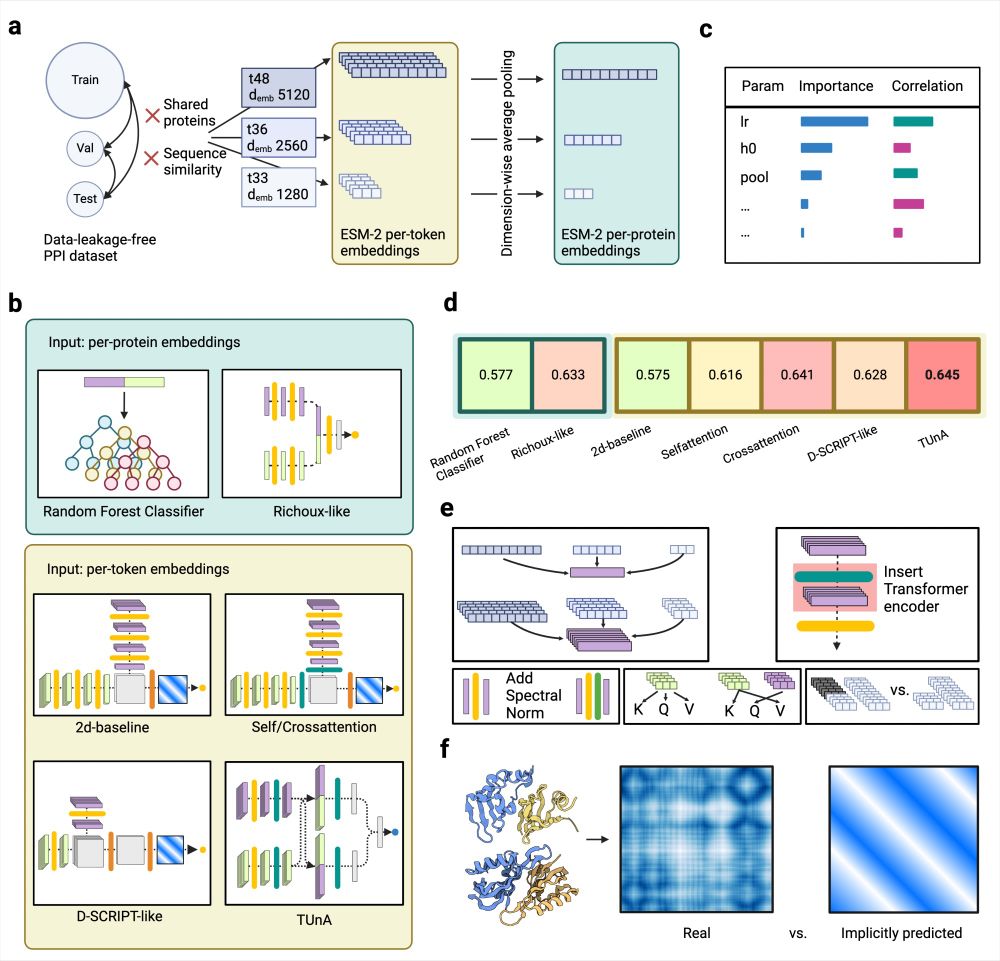
Graphical summary of the analyses done in the publication displayed on six panels a-f. (a) We computed ESM-2 embeddings of different sizes for the proteins of our data-leakage-free PPI dataset. The per-token embeddings have variable sizes depending on the protein length, while the per-protein embeddings have a fixed size by applying dimension-wise averaging. (b) We tested two models operating on the per-protein embeddings—a baseline random forest classifier and adaptions of the previously published Richoux model. Five models operated on the per-token embeddings: a 2d-baseline, the 2d-Selfattention and 2d-Crossattention models (which expanded the 2d-baseline through a Transformer encoder), and adaptations of the published models D-SCRIPT and TUnA. (c) Hyperparameter tuning gave us insight into the influence of each tunable parameter on the classification performance. (d) No model surpassed an accuracy of 0.65. The more advanced models had similar accuracies, leading us to believe that the information content of the ESM-2 embedding has more influence than the model architecture. Per-token models did not consistently outperform per-protein models. (e) We applied various modifications to test their influence: different embedding sizes, inserting a Transformer encoder into different positions, adding spectral normalization after the linear layers, self- vs. cross-attention, and removing the padding. (f) Finally, we compared the implicitly predicted distance maps of the 2d-baseline, 2d-Selfattention, 2dCrossattention, and D-SCRIPT-ESM-2 to real distance maps computed from PDB structures.
🧬🖥️ Proud to share our latest update on PPI predictions – "Deep learning models for unbiased sequence-based PPI prediction plateau at an accuracy of 0.65" doi.org/10.1101/2025... by T. Reim, published with @itisalist.bsky.social @dbblumenthal.bsky.social, A. Hartebrodt, and me. What did we do? 1/15 🧵
27.01.2025 10:58 — 👍 10 🔁 3 💬 1 📌 2

Emergence of power-law distributions in protein-protein interaction networks through study bias
Incredibly happy to finally see our manuscript "Emergence of power-law distributions in protein-protein interaction networks through study bias" published in @elife.bsky.social. doi.org/10.7554/eLif... It's been a long but fun journey with @dbblumenthal.bsky.social and @martinschaefer.bsky.social
14.12.2024 19:47 — 👍 16 🔁 9 💬 1 📌 1
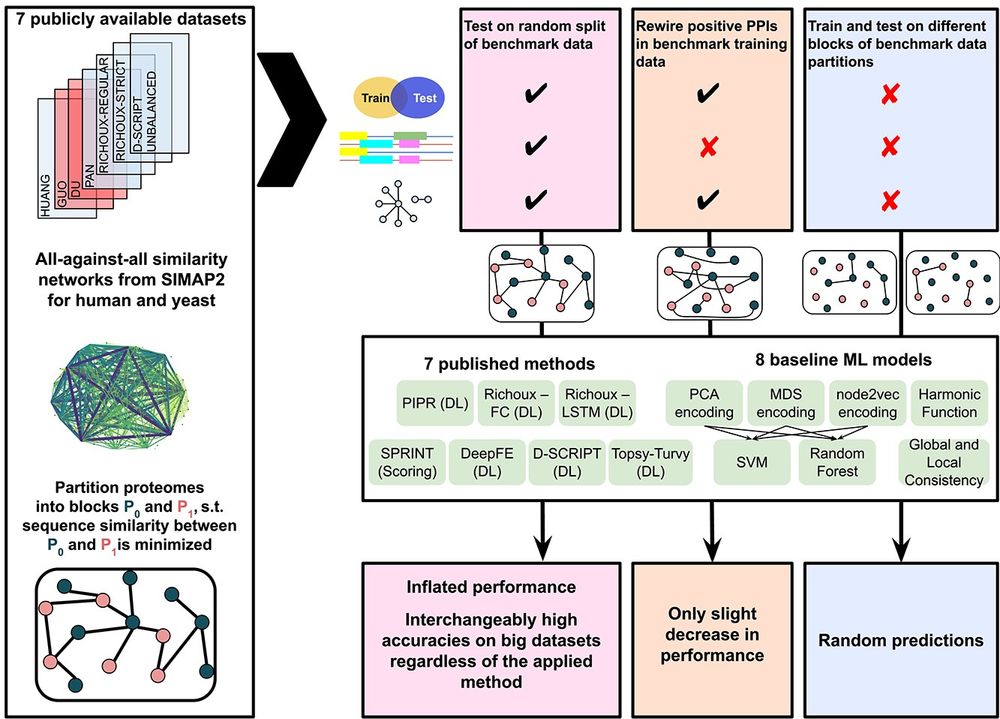
Overview figure summarizing the major analyses of the paper. In the setting employed by published methods, performances are strongly inflated regardless of model and dataset. When the positive training dataset is randomly rewired, there is only a slight decrease in performance. When the models are evaluated on unseen proteins that are unsimilar to the training proteins, performances become random.
🧬💻Transferring my paper posts here, starting off with "Cracking the black box of deep sequence-based protein-protein interaction prediction" doi.org/10.1093/bib/..., which I published together with @itisalist.bsky.social and @dbblumenthal.bsky.social. So what was it about? 1/13 🧵
28.11.2024 09:10 — 👍 14 🔁 4 💬 3 📌 0
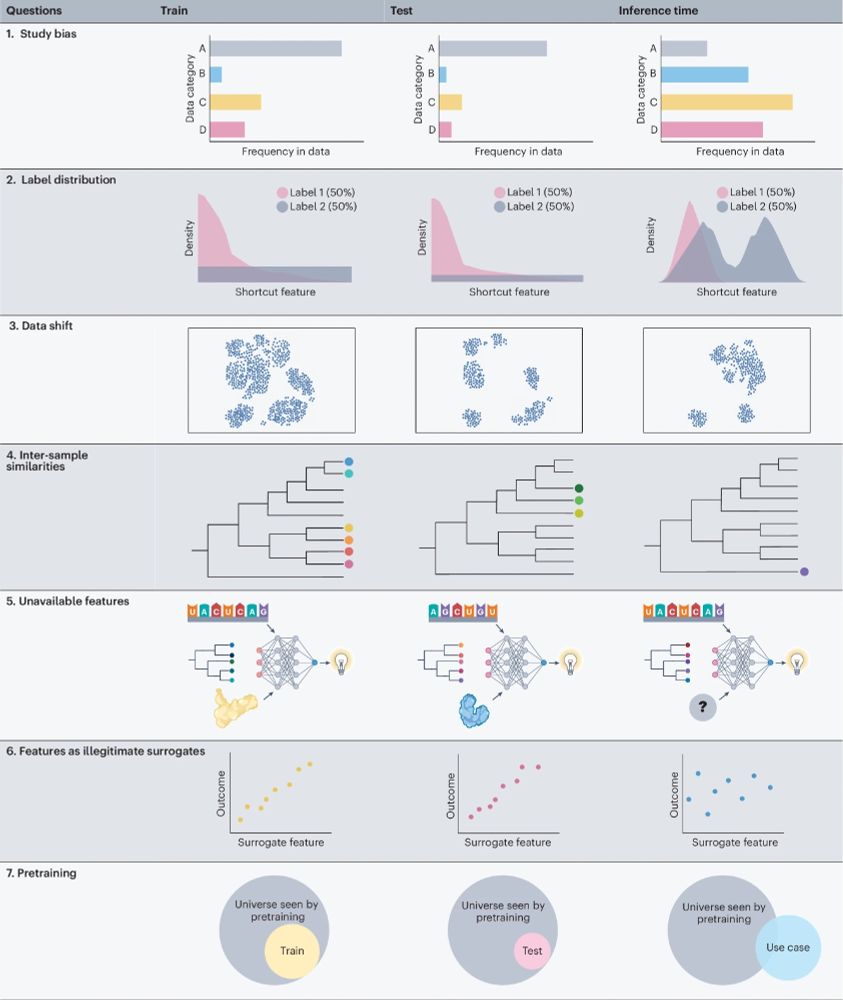
Overview figure of the seven guiding questions addressed in manuscript regarding (1) Study bias, (2) Label distribution, (3) Data shift, (4) Inter-sample similarities, (5) Unavailable features, (6) Features as illegitimate surrogates, (7) Pretraining
🧬🖥️Tranferring my papers, Pt. 2: "Guiding questions to avoid data leakage in biological machine learning applications" doi.org/10.1038/s41592-024-02362-y, which I published with @itisalist.bsky.social @dbblumenthal.bsky.social @romanjoeres.bsky.social, D. Grimm, F. Haselbeck, and O.V. Kalinina. 🧵1/20
02.12.2024 11:53 — 👍 6 🔁 2 💬 1 📌 0
Reminder: #computational_biology position available in my team:
- large-scale #proteomics #metabolomics data analysis
- #rstats , #python, #machinelearning
- great living in the ❤️ of the 🇮🇹 Alps ⛰️
👉 info and apply: bit.ly/3wztFsN
01.03.2024 07:53 — 👍 2 🔁 2 💬 0 📌 1
PhD student in bioinformatics and ML, Tartu University 🎓
proud Ukrainian 🇺🇦
mountain nerd 🤓
Data Science in Systems Biology: We are a research group at the TUM School of Life Sciences. Cutting-edge expertise is united here in order to unlock the mechanisms of various systems in the human body.
🧬 Structural Bioinformatics | 💊 AI/ML for Drug Discovery | Geometric DL
🔬 @iocbprague.bsky.social, prev. PhD @cusbg.bsky.social @mff.unikarlova.cuni.cz
Professor of Phytopathology | Kiel University
| Crop wild relatives and their pathogens | Likes quantitative and qualitative resistance mechanisms & popgen
Assistant Professor leading the Computational Biomedicine Group at the University of Innsbruck • Data scientist at the Digital Science Center (DiSC) • Precision medicine and cancer immunotherapy • Mother 🧬🖥️
https://computationalbiomedicinegroup.github.io/
Bioinformatics PhD student at TUM 🧬🖥️ 👩💻 Currently interested in machine learning pitfalls, drug response prediction, and protein-protein interaction prediction.
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
Quantitative Single-Cell Biology
Prof at Würzburg Institute of Systems Immunology (WüSI)
Decoding tumor (epi)genomes @ IEO in Milan | https://www.schaeferlab.org/
Assistant Professor for Data Science in Systems Biology at the Technical University of Munich (http://daisybio.de). Mostly posting about bioinformatics and systems / network biology research. Views are my own. he / him.
Official account of the Friedrich-Alexander-University Erlangen-Nürnberg (FAU). We are Moving Knowledge.
fau.de
Impressum: https://www.fau.de/impressum/
Datenschutz: https://www.fau.de/datenschutz/
Wir ermutigen, beraten und fördern Initiativen gegen Rechtsextremismus, Rassismus und Antisemitismus!
Spenden: https://www.amadeu-antonio-stiftung.de/spenden-und-stiften/
Datenschutz: https://www.amadeu-antonio-stiftung.de/datenschutz/
Esoterik-Kritik.
Blogger, Podcaster, Journalist. Antifaschist, Antitheist, Queer.
„Nur weil ein Garten schön ist, muss man nicht daran glauben, dass auch Feen darin hausen.“ (Douglas Adams)
https://Anthroposophie.blog
Journalist, Moderator, Fragensteller. Bildung, Wissenschaft, Beruf
Das deutschsprachige @RealScientists.
Echte Wissenschaft von echten ForscherInnen, AutorInnen, KommunikatorInnen, KünstlerInnen... Diese Woche: @sarahbusch.bsky.social
Institute of Medical Informatics, University of Münster
Postdoctoral researcher at @anshulkundaje.bsky.social Machine learning #ML, gene regulatory networks #GRN, single cell and spatial #omics.
Previously at @saezlab.bsky.social
Core developer of https://decoupler.readthedocs.io/ at @scverse.bsky.social
Computer scientist, currently president of FAU Erlangen-Nürnberg.
🇪🇺 // JOIN ME ON MASTODON: https://edi.social/@janboehm // @ZDF Magazin Royale 🦠/ fest & flauschig @Spotify / hotline +49 30 959997666 / ❤️✊️😉 / unvernunft.berlin / boehmermann.de






