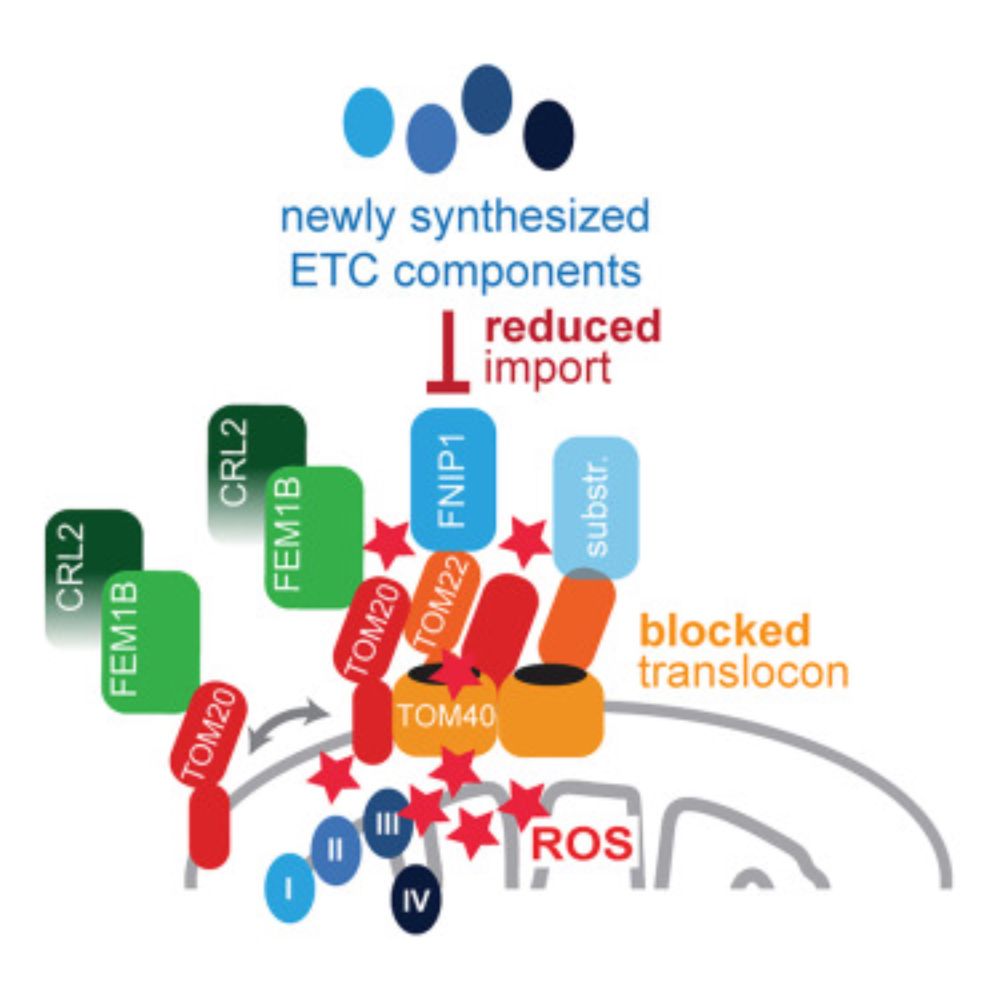
A conserved mechanism for the retrieval of polyubiquitinated proteins from cilia
The temporospatial distribution of proteins within cilia is regulated by intraflagellar transport (IFT), wherein molecular trains shuttle between the cell body and cilium. Defects in this process impair various signal-transduction pathways and cause ciliopathies. Although K63-linked ubiquitination appears to trigger protein export from cilia, the mechanisms coupling polyubiquitinated proteins to IFT remain unclear. Using a multidisciplinary approach, we demonstrate that a complex of CFAP36, a conserved ciliary protein of previously unknown function, and ARL3, a GTPase involved in ciliary import, binds polyubiquitinated proteins and links them to retrograde IFT trains. CFAP36 uses a coincidence detection mechanism to simultaneously bind two IFT subunits accessible only in retrograde trains. Depleting CFAP36 accumulates K63-linked ubiquitin in cilia and disrupts Hedgehog signaling, a pathway reliant on the retrieval of ubiquitinated receptors. These findings advance our understanding of ubiquitin-mediated protein transport and ciliary homeostasis, and demonstrate how structural changes in IFT trains achieve cargo selectivity. ### Competing Interest Statement The authors have declared no competing interest. Sara Elizabeth O'Brien Trust Postdoctoral Fellowship awarded through the Charles A. King Trust Postdoctoral Research Fellowship Program, , 8460873-01 Richard and Susan Smith Family Foundation, https://ror.org/05j95n956, National Institute of General Medical Sciences (NIGMS), , R01GM141109, R01GM143183
How do cells keep their cilia “clean” and functional? Our new study uncovers a conserved mechanism for retrieving polyubiquitinated proteins from #cilia – a process essential for cellular signaling and health. #cellbiology #ciliopathy #ubiquitin #IFT 🧵👇 1/n
29.04.2025 15:04 — 👍 53 🔁 26 💬 2 📌 2
Well done, Sven! It’s in my to-read list for the week!
29.04.2025 19:14 — 👍 2 🔁 0 💬 0 📌 0
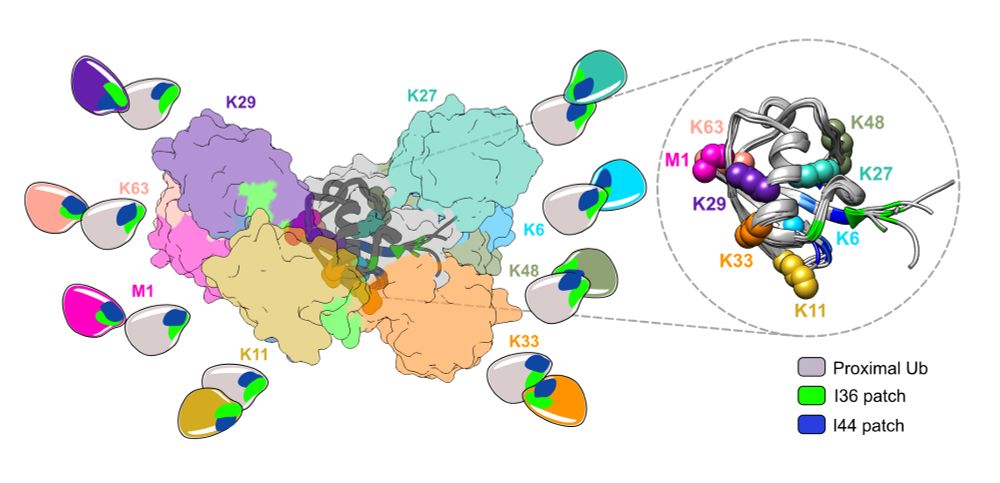
'Ubiquitin Lysine-Linkages' from the review 'Ubiquitin: a structural perspective' in Molecular Cell
New to #ubiquitin? Been with it for a while? Either way this review may be for you 🤩
Excited to share our review article in @molcell.bsky.social, diving deep into everything #ubiquitin
Read here 👉: kwnsfk27.r.eu-west-1.awstrack.me/L0/https:%2F...
@cellpress.bsky.social
@wehi-research.bsky.social
17.01.2025 02:19 — 👍 56 🔁 23 💬 1 📌 0

Nikita Sergejevs
Read more about this research in our ‘First person’ interview with Nikita Sergejevs: journals.biologists.com/jcs/article/...
18.12.2024 15:05 — 👍 8 🔁 1 💬 0 📌 0
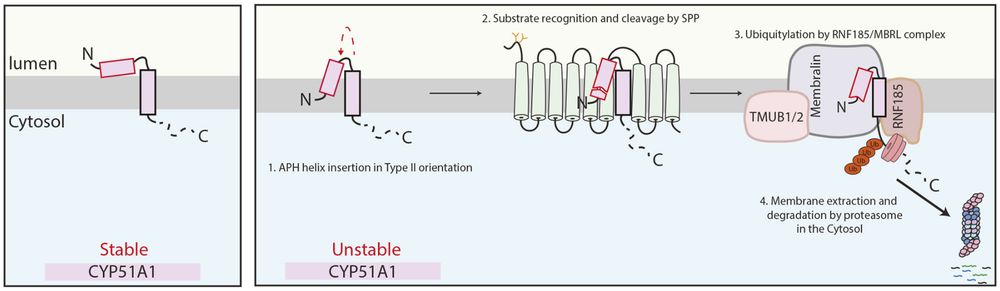
Proposed model of CYP51A1 recognition by SPP. In cells, the lanosterol demethylase CYP51A1 can exist in two distinct conformations, with only one conformation being recognized for membrane protein quality control. The early recognition event by SPP consists of an N-terminal cleavage within the AH of the substrate. This generates a truncated version of CYP51A1 that is subsequently recognized by the RNF185–MBRL E3 ubiquitin ligase complex, extracted from the ER membrane and degraded by the cytosolic proteasomes. APH, amphipathic helix.
Nikita Sergejevs, Pedro Carvalho and colleagues find that that the intramembrane protease SPP conducts topology surveillance of CYP51A1 by initiating quality control of incorrectly inserted molecules.
Highlight: journals.biologists.com/jcs/article/...
Article: journals.biologists.com/jcs/article/...
18.12.2024 15:05 — 👍 13 🔁 5 💬 1 📌 1
Congratulations! Lucky us in Dundee as we will get to hear your exciting research, past and present when you deliver the Adam Neville Lecture tomorrow. I am very much looking forward to it!
12.12.2024 21:56 — 👍 1 🔁 0 💬 0 📌 0

Graphic Abstract for "Yeast Knowledge Graphs Database for Exploring Saccharomyces cerevisiae and Schizosaccharomyces pombe"
🚨 Our latest study, "Yeast Knowledge Graphs Database for Exploring Saccharomyces cerevisiae and Schizosaccharomyces pombe," has just been deposited in @biorxivpreprint.bsky.social ! We’re thrilled to share our work with the world! #KnowledgeGraphs
www.biorxiv.org/content/10.1101/2024.12.04.626523v1
10.12.2024 04:24 — 👍 31 🔁 9 💬 2 📌 3
Waves of new BlueSky arrivals in last few days!
I’ve been tracking starter packs relevant to molecular/mechanistic/genetic/cellular…etc biology
Hope it’s helpful but warning: now a v long thread!
Probably the last time I can do this...
12.11.2024 15:03 — 👍 194 🔁 115 💬 16 📌 19
If you are interested in doing cutting-edge molecular #neurobiology research, I can highly recommend the opportunity to work with @adamgrieve.bsky.social, whom not only will ensure you an exciting scientific journey, but will also be a great mentor, guiding you through the ups and downs of a #PhD.
08.12.2024 21:20 — 👍 3 🔁 0 💬 1 📌 0
Amazing PhD opportunity to join an exciting new lab at Bristol Biochemistry - send your best students in Ferdos’ direction!
27.11.2024 12:36 — 👍 3 🔁 1 💬 1 📌 0
This study has it all! A truly structure-to-function study on two #deubiquitinases that were thought to be pseudoenzymes and providing insights into disease mutations. Congratulations to @gallantkai.bsky.social, Kim and the team!
06.12.2024 19:36 — 👍 2 🔁 0 💬 1 📌 0
Since I read about AKIRIN2 in 2021 in their earlier paper in Nature, I have always been fascinated by this protein. This follow-up preprint is aimed at unraveling the mechanistic aspects of its functions! Just excited at the fact that we have so much more to learn about the world of #proteostasis!
06.12.2024 19:20 — 👍 4 🔁 2 💬 0 📌 0
It was a treat to hear this story when @micharapelab.bsky.social visited University of Dundee and continuation of the story at the EMBO #ubiquitin and #ubiquitin-like proteins meeting in Cavtat, a couple months ago! Very elegant and informative mechanistic and functional works on #stresssignalling!
06.12.2024 19:08 — 👍 3 🔁 0 💬 1 📌 0

SAVE THE DATE 🗓️Ubiquitin & Friends Symposium: 8-9 May 2025 in Vienna!
Registration will open🔜 Keep an eye out for the announcement 👀
✨guest speaker lineup👇🧵 +talks from abstracts! Great chance for #ECRs to meet with peers & experts in a friendly setting!
🔗 www.protein-degradation.org/symposium/
29.11.2024 14:18 — 👍 59 🔁 28 💬 1 📌 3
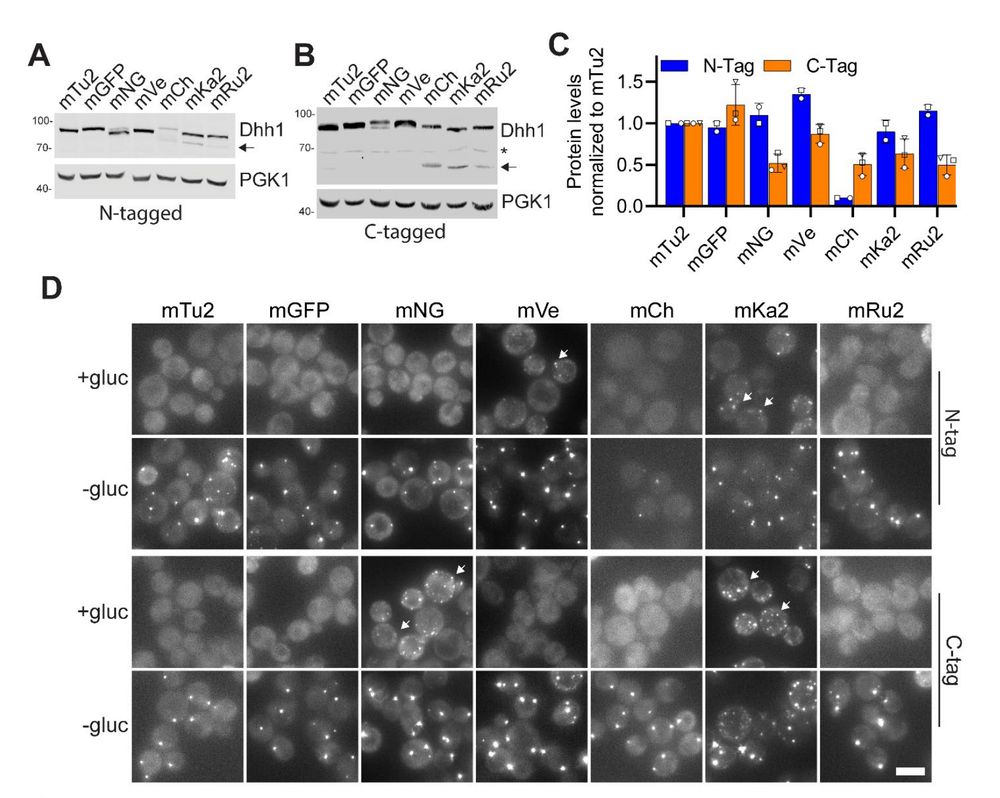
Figure 4. Position and type of fluorescent tag affect protein expression and P-body numbers in vivo.
The dark side of fluorescent protein tagging: the impact of protein tags on biomolecular condensation
www.biorxiv.org/content/10.1...
25.11.2024 12:19 — 👍 306 🔁 119 💬 10 📌 7
Cellular and protein homeostasis webinars - EPFL - Ecole polytechnique fédérale de Lausanne
For people interested in chaperones, protein quality control, proteostasis and cellular homeostasis, here is the link of the webinars we (P. Goloubinoff, A. Barducci, N. Nillegoda and I) hosted during the COVID pandemics. A lot of material!
tinyurl.com/27y5wpzy
17.11.2024 12:18 — 👍 26 🔁 13 💬 2 📌 2
Hello! As a new user, I thought I would introduce myself. I am a postdoc in the lab of @kulathu.bsky.social at MRC-PPU, Dundee, UK. Fascinated by #molecularchaperones and #proteinqualitycontrol. Find me on pubmed pubmed.ncbi.nlm.nih.gov?term=Logesva... or here scholar.google.com/citations?us...
18.11.2024 02:06 — 👍 10 🔁 1 💬 0 📌 0
Hi Dorien, Loges here from Dundee, UK. Postdoc, a molecular cell biologist. I would really like to be added to this group. Thank you!
18.11.2024 01:57 — 👍 1 🔁 0 💬 0 📌 0

We are still considering postdoc candidates for the post advertised below. A 2-year contract beginning in 2025 to work on protein SUMO E3 ligases and the nucleosome.
16.11.2024 06:43 — 👍 14 🔁 8 💬 0 📌 5
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
Sendoel lab at the University of Zurich, alumnus of the The Rockefeller University, NYC.
We are excited about mRNA translation, stem cells and cancer.
www.sendoellab.org
Postdoc researcher at the Helmholtz Diabetes Center in the @nataliekrahmer.bsky.social Lab and the Perocchi Lab | Formerly University of Oxford | (she/her)
The Bsky account for the UKRI funded Foldamer Network: foldnet.uk
Reader (Associate Professor) at Imperial College London
Home of mitochondrial transport proteins
Some days you’re a fish exploring new waters. Other days, you’re stuck in a maze making the same turn over and over. Either way, keep swimming.
Neuroscientist and pharmacologist
PhD Student in the lab of Tim Clausen
C. elegans
Muscle function
Starvation
mTORC1/2
Vienna
Assist. Prof. in Critical Mineral Resources at the University of Calgary | Science & Policy | GeoLatinas | 🇨🇴🇳🇱🇨🇦 | she/her/ella
Posts in English & Español
Scientist and Deputy Editor, Cell at Cell Press
All things science with a love for #microbes, #immunology & #metabolism #sciencesky #womeninSTEM - Opinions and posts are my own. 🇺🇸🇮🇳
https://www.cell.com/cell/editors-and-staff
Professor of Theoretical Chemistry @sorbonne-universite.fr & Head @lct-umr7616.bsky.social| Guest Faculty @ Argonne| Co-Founder & CSO @qubit-pharma.bsky.social (My Views) |
https://piquemalresearch.com | https://tinker-hp.org
Research fellow at Aston Institute of Membrane Excellence. Previously folding proteins at The Crick and membrane proteins at King's College London.
https://isb.med.upenn.edu/
Group leader in membrane synthetic biology at ANU, Ngunnawal country. I like cryo-EM, yeast, biosensors and riding bikes. he/him/his.
Ad Astra Fellow at UCD, formerly Henry Dale and Lister Institute Senior Research Fellow at the University of Sheffield
Scientist, LNP Services - Echelon Biosciences
Here for Molecular Biology, RNA, Lipids, and any relevant spirited discussion.
🇲🇾 scientist in 🇦🇺 | 🦟 🩸 #malaria |🦙 #nanobodies | Prof @WEHI_research @ourANU | on Wurundjeri, Ngunnawal & Ngambri lands | views my own 🏃🏻♀️ 🍜
Mechanosensitive channels & Membrane Proteins - Structural EPR Spectroscopy at The University of Manchester - DEER/PELDOR/PDS
pliotasgroup.org
Doing drug discovery things. Senior Scientist at Schrodinger. Views are my own.