Very excited about this work by @mrazo.bsky.social in collaboration with www.madhavmani.com bsky.app/profile/mraz... I am so lucky to be able to collaborate with such brilliant people! Learned a lot. This is our first foray into ML approaches and I am quite taken with their power 1/n
15.05.2025 18:16 — 👍 26 🔁 9 💬 1 📌 0
26/n Thank you for reading. If you have any questions or comments, please reach out!
15.05.2025 14:33 — 👍 4 🔁 0 💬 0 📌 0
25/n We're excited to see how this approach can be applied to other problems in evolutionary biology and beyond
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
Learning the Shape of Evolutionary Landscapes: Geometric Deep Learning Reveals Hidden Structure in Phenotype-to-Fitness Maps
24/n Furthermore, we have a custom website for the paper, where you can find the HTML version as well as detailed notebooks explaining the computational approaches involved in the project. Shout-out to @quarto.org for facilitating the creation of this website!
mrazomej.github.io/antibiotic_l...
15.05.2025 14:33 — 👍 1 🔁 0 💬 2 📌 0
GitHub - mrazomej/antibiotic_landscape: Repository for the exploration of the fitness landscape of antibiotic resistance.
Repository for the exploration of the fitness landscape of antibiotic resistance. - mrazomej/antibiotic_landscape
23/n This work bridges evolutionary biology and machine learning, showing how geometry-aware deep learning can reveal hidden structure in biological complexity. All of the @julialang.org code utilized for this project is available at github.com/mrazomej/ant...! #OpenScience
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
22/n The broader implication: evolution may be more predictable than we thought, with organisms following constrained paths as they adapt to new environments.
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
21/n This approach could help us better understand and predict evolutionary trajectories, with potential applications for antibiotic resistance, viral evolution, and cancer treatment.
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
20/n As with the simulated data, our 2D nonlinear representation worked better than linear methods (like PCA) with a significantly larger number of dimensions. Simpler AND more accurate! #DataScience
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
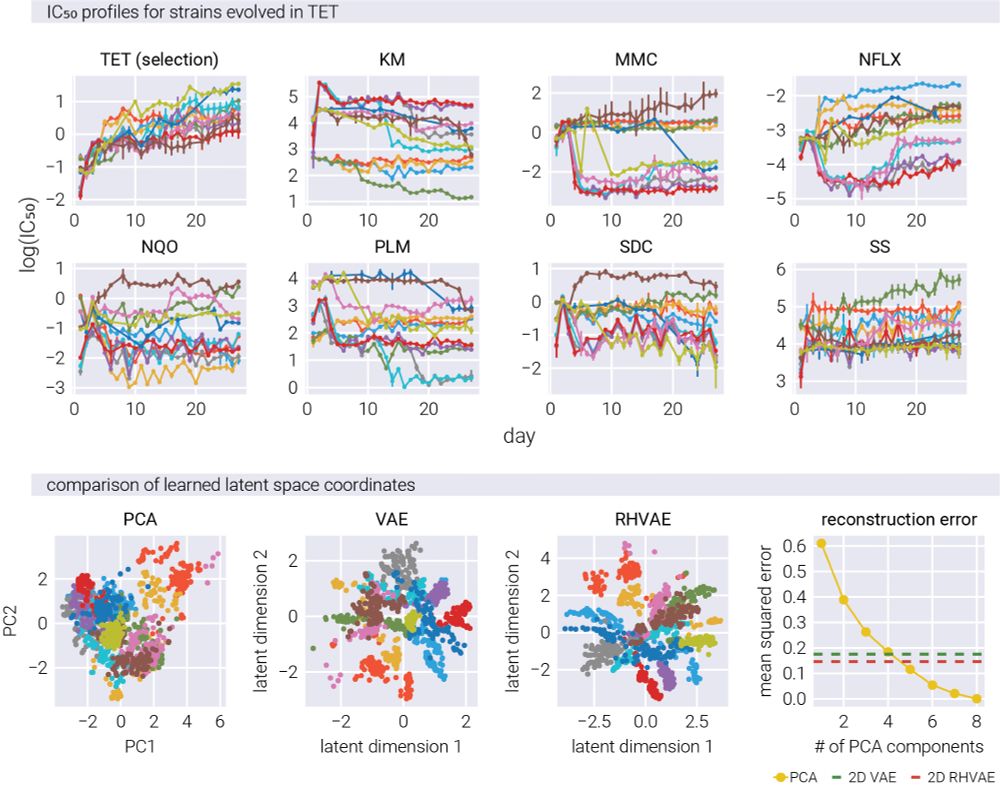
19/n The results? We can represent complex resistance patterns in just two dimensions while preserving key relationships!
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
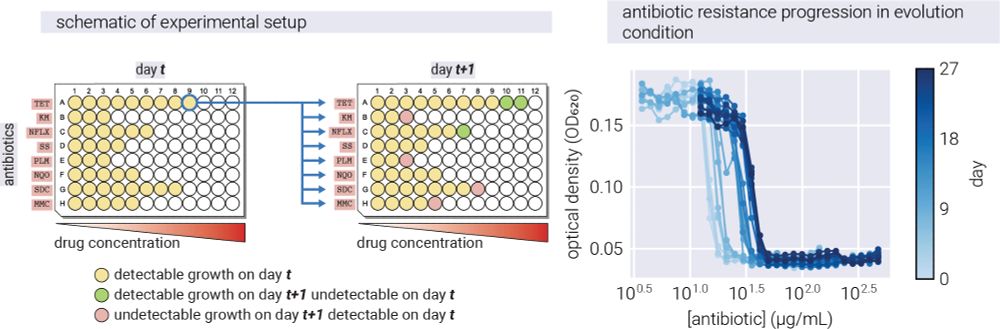
18/n Then we applied it to real-world data: E. coli evolving under different antibiotics. For this, we used the data from the incredible paper by Iwasawa et al. 2022, where they measured the fitness of E. coli evolving under different antibiotics.
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
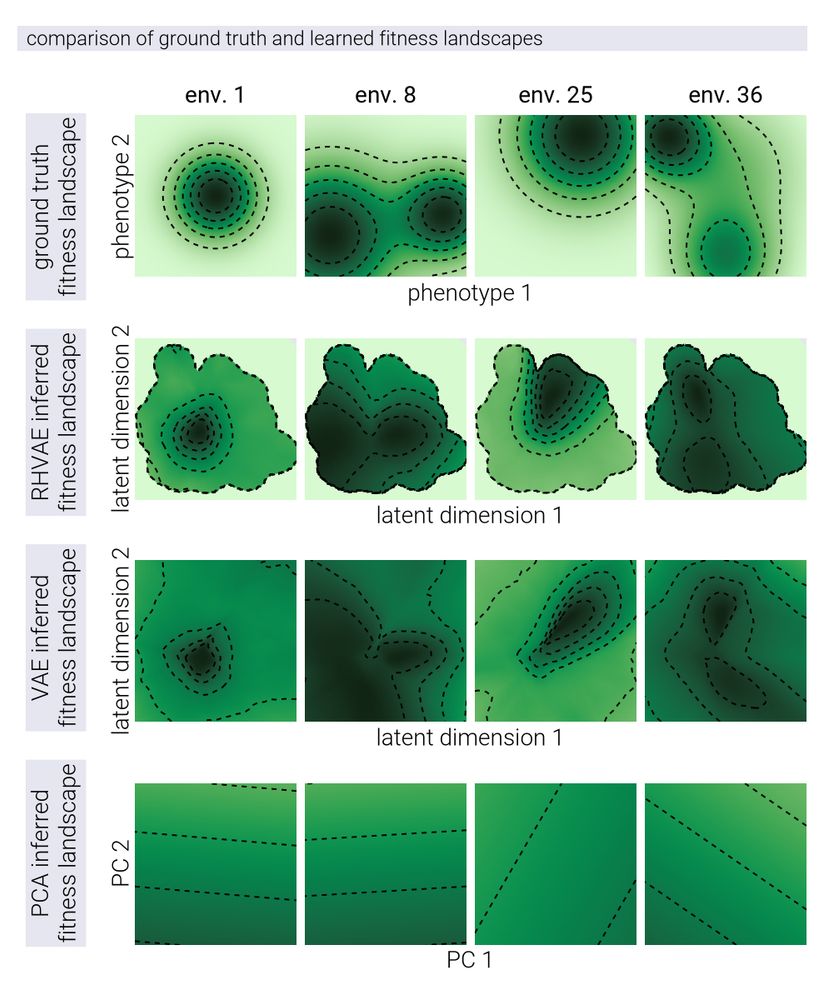
17/n Another cool feature of the RHVAE applied to this problem is that it allowed us to qualitatively reconstruct the underlying fitness landscapes from which the adaptive walks were drawn.
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
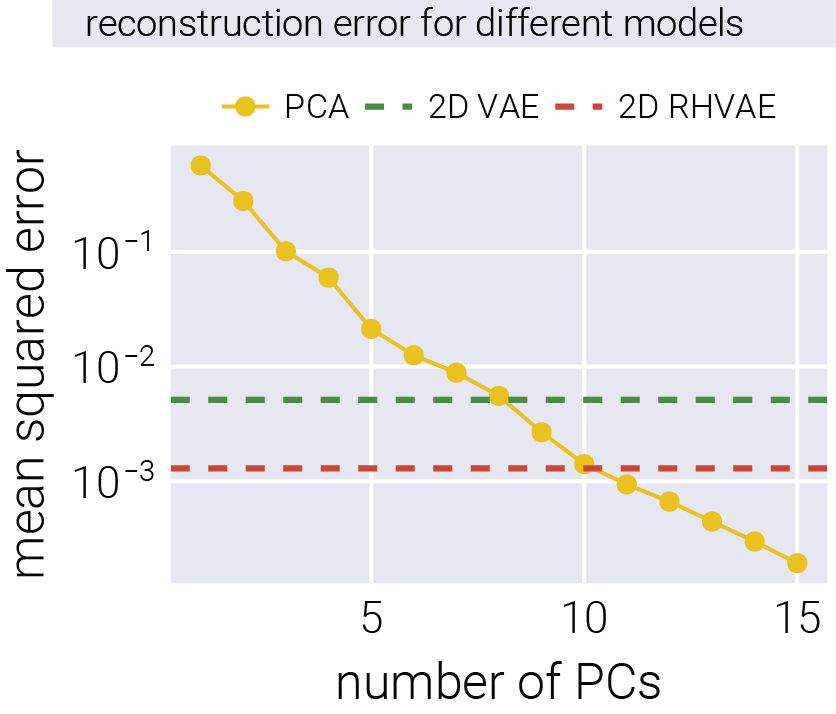
16/n Moreover, with only two non-linear dimensions, the RHVAE has the same reconstruction accuracy as a 10-dimensional PCA!
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
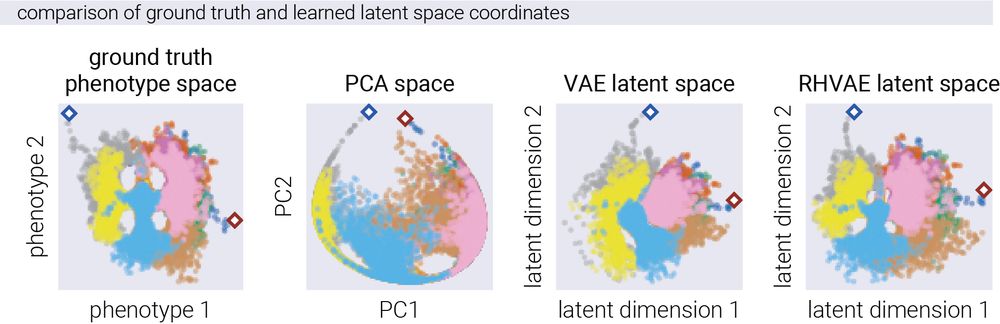
15/n For this, we compared the performance of a linear model (PCA), a vanilla variational autoencoder (VAE), and our RHVAE. The non-linear models accurately reconstructed the underlying structure and relationships that generated the fitness patterns.
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
14/n From this data, we can then fit a model to reconstruct the phenotypic coordinates of each genotype only from the fitness data. In other words, given that we only get the "z-axis" in multiple environments, we ask: can we recover the relative "x" and "y" coordinates of each genotype?
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
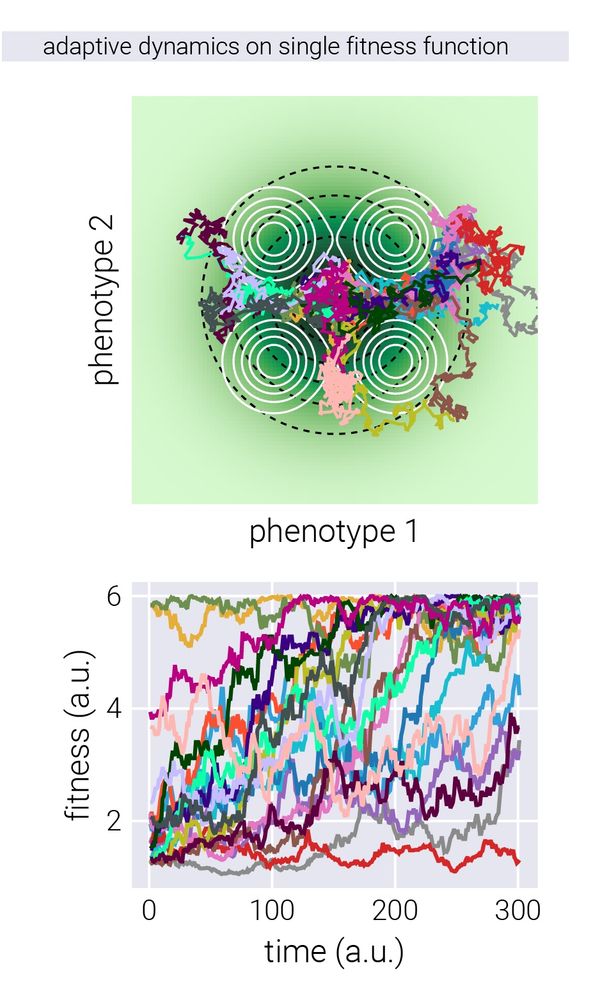
13/n Given this picture, we can then simulate adaptive walks in this phenotype space, and measure the fitness of the genotypes along the way.
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
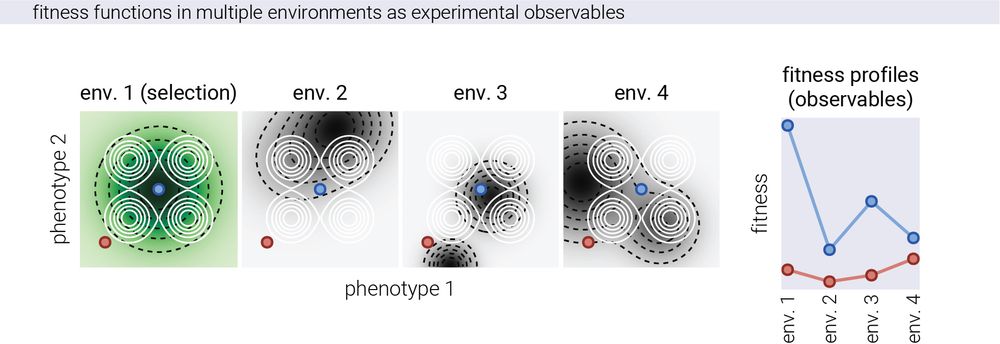
12/n We first tested this on simulated data. For this, we took the conceptual picture of Fisher’s geometric model seriously and imagined organisms with fixed coordinates in phenotype space, while the fitness landscape defined by the environment changes, giving different fitness readouts.
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
11/n Think of it like creating a 2D map of Earth's 3D surface, but also knowing exactly how distances on the map relate to real distances anywhere on the planet. That's what our RHVAE does with complex fitness data!
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
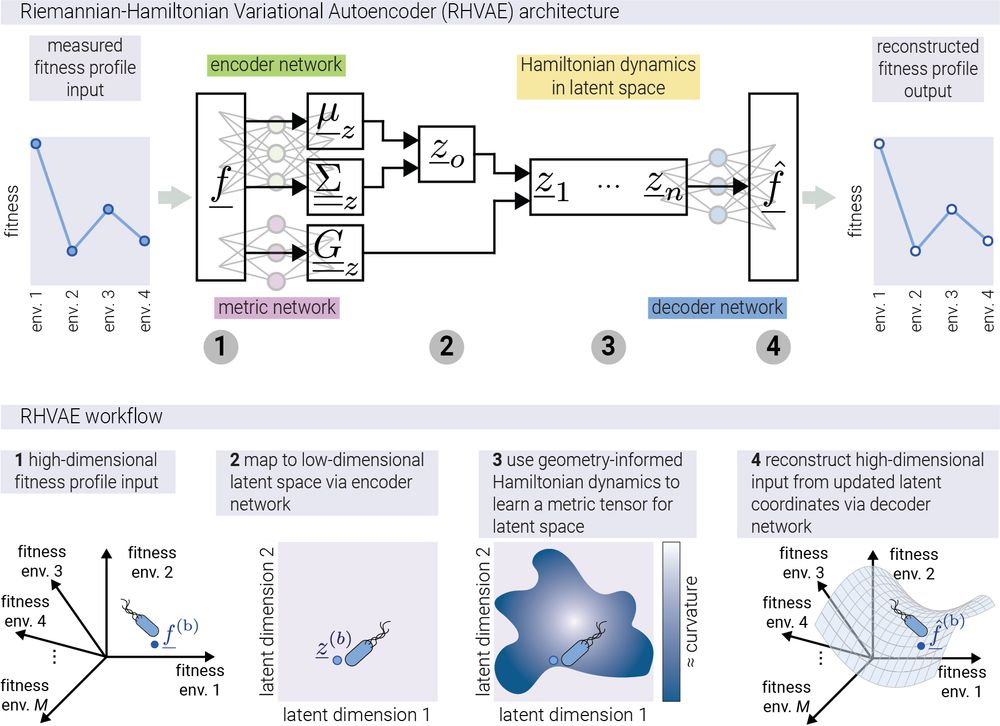
10/n To relax this assumption, we take advantage of the progress in geometric deep learning. More specifically, we use a neural network called a "Riemannian Hamiltonian Variational Autoencoder" (RHVAE) that not only reduces dimensionality but preserves the geometric relationships between data points
15.05.2025 14:33 — 👍 4 🔁 1 💬 1 📌 0
9/n This strong assumption can limit our ability to uncover the true dimensionality of the adaptive phenotypic landscape as phenotypes could be non-linearly related to fitness. This is where our paper comes in!
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
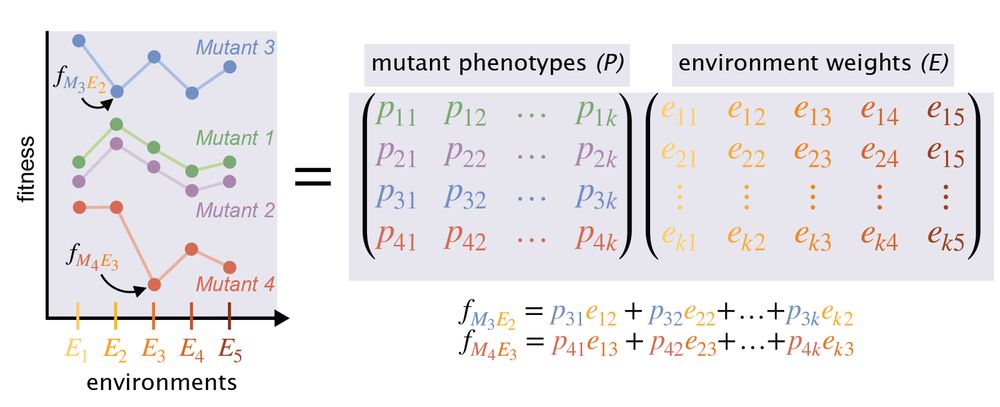
8/n This approach involved a linear decomposition of the fitness matrix via SVD. In other words, the authors assumed that fitness is a linear function of the phenotypic features.
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
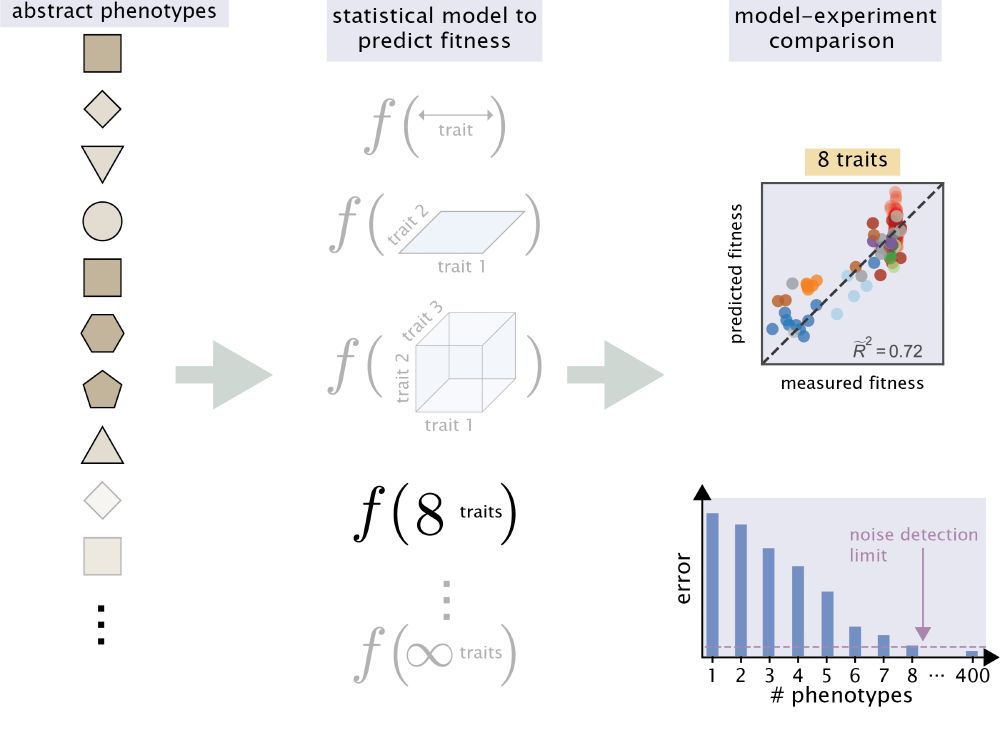
7/n The task is then to use a statistical model that takes as input some abstract phenotypic features and predicts the fitness of the genotype. In this way, Kinsler et al. 2020 found that 8 of these features were sufficient to predict their data
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
6/n The idea being that the GxE (genotype-environment) variation in fitness must be due to the phenotypic effects and thus the structure of the GxE variation in fitness can be used to infer phenotypic layer without measuring any phenotypes explicitly
15.05.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
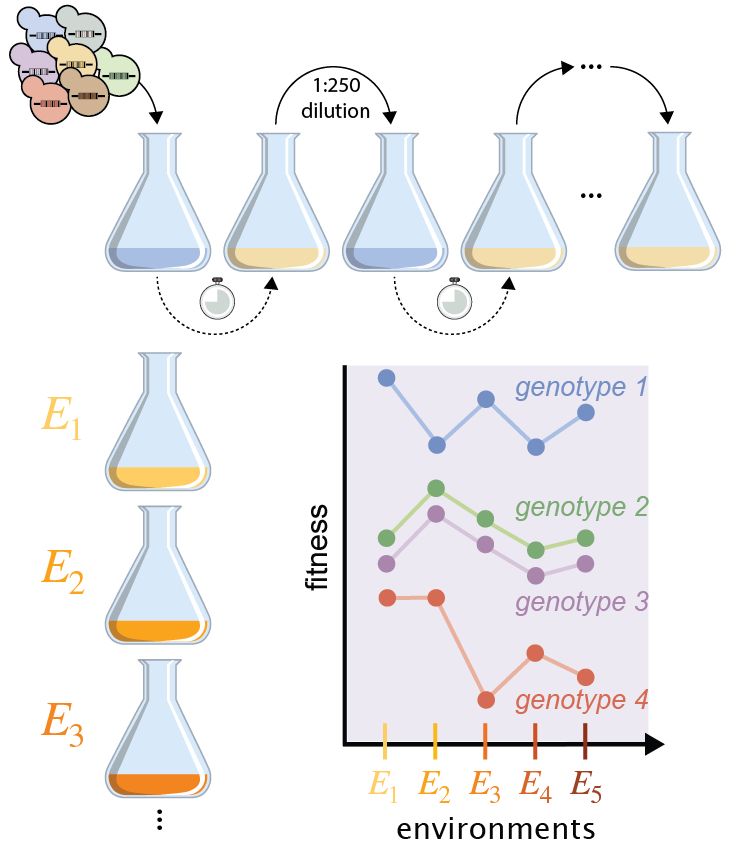
5/n Our lab and others have taken an approach based on our ability to measure fitness in the lab for multiple genotypes in different environments. For example, Kinsler et al. 2020 & Gosh et al. 2025 determined the fitness of many yeast genotypes in different environments.
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
4/n In other words, given that we are able to track a microbial population as it evolves in the lab, are there a limited set of phenotypic changes that are frequent enough and adaptive enough (large µs in the pop gen lingo) that keep appearing in the population over and over again?
15.05.2025 14:33 — 👍 2 🔁 0 💬 1 📌 0
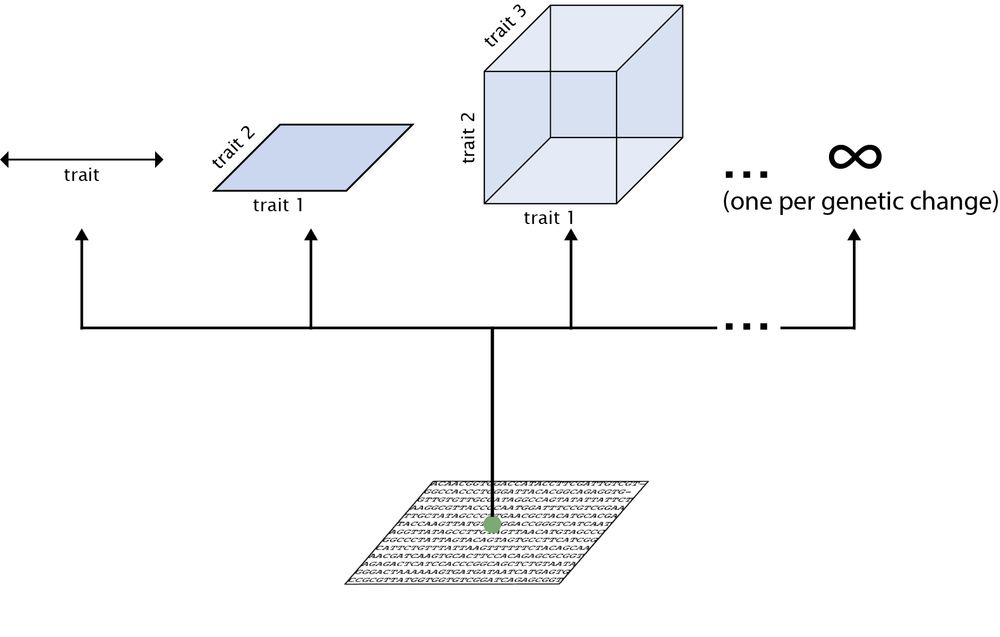
3/n A simpler question one can ask is: when populations adapts to an environment, are there a limited set of phenotypic changes that dominate the adaptive process? I.e., can we uncover the dimensionality of the adaptive phenotypic landscape we observe in experimental evolution setups?
15.05.2025 14:33 — 👍 6 🔁 0 💬 1 📌 0
2/n A central challenge in evolutionary biology: Can we predict how organisms adapt to different environments? The sheer number of possible genetic changes and the lack of understanding of the underlying phenotypic changes driving adaptation makes this incredibly difficult to answer.
15.05.2025 14:33 — 👍 5 🔁 0 💬 1 📌 0
1/n 🧵 Excited to share our new paper! We developed a framework to reveal hidden simplicity in how organisms adapt to different environments, particularly focusing on antibiotic resistance evolution. #EvolutionaryBiology #MachineLearning
15.05.2025 14:33 — 👍 37 🔁 22 💬 1 📌 1
06.12.2024 14:54 — 👍 0 🔁 0 💬 0 📌 0
I am a New Yorker staff writer and author of "H is for Hope," "Under a White Sky," and "The Sixth Extinction," and forthcoming “Life on a Little-Known Planet”, out in November. Learn more at elizabethkolbert.com.
Single cell systems and synthetic biology lab at Northwestern University and Chan Zuckerberg Biohub Chicago.
https://www.goyallab.org/
Biophysicist working at the edge of single cell biology, machine learning, and statistical-mechanics. Postdoc in Yogesh Goyal's lab at Northwestern; PhD in biophysics, UC Berkeley; BS in math and physics, CWRU.
Assistant professor in Cell and Developmental Biology at UCSD
@ucsandiego.bsky.social. Loves biophysics, cell mechanics and motility, parasite biology.
@embl.org PhD & Bridging Excellence Postdoc (LifeScienceAlliance) with
@petrovadmitri.bsky.social @stanford.edu and Aulehla & Steinmetz labs @embl.org using #SystemsEvolution to study #Aging
Full-time father, part-time postdoc at Thornton Lab at UChicago | Evolutionary genetics
Postdoc @ Harvard with Michael Desai | Evolutionary dynamics
ocf.berkeley.edu/~joaoascensao
Microbial evolutionary biologist at Stanford for most of the time. Radically queer liberal for the rest. 🇪🇺🏳️🌈 They/Them
Postdoc at Stanford University.
Interested in pop. gen., adaptation, and a wholesome scientific community.
he/him
Assistant Professor of Physics at WashU working in areas of biophysics and soft matter.
PostDoc University College London 📚 | ecology and evolution of microbes 🦠 | 🇲🇽🇬🇧 | romanzapien.github.io
Researcher in machine learning
Interested in #physics of life. Theorist but some of my best friends are experimentalists.
Professor of Marketing at NYU Stern School of Business, serial entrepreneur, and host of the Prof G and Pivot Podcasts.
Tidier.jl is a data analysis package inspired by R's tidyverse and crafted specifically for Julia.
https://tidierorg.github.io/Tidier.jl/dev/
Emergence, Networks & Complexity | Collective Behavior(s) from Cells to Societies 🧬🦠🧠🌇
Prof. @UniPadova, Galileo's University | Lab: @comunelab.bsky.social | Web: https://linktr.ee/manlius
Thoughts at manlius.substack.com
Theorist at @normalcomputing.com. Thermodynamic computing for efficient AI.
Python library for creating static, animated, & interactive visualizations. Chat w/ us @ https://discourse.matplotlib.org/
Sponsored by NumFocus
At KITP on the UC Santa Barbara campus, researchers in theoretical physics and allied fields collaborate on questions at the leading edges of science.
www.kitp.ucsb.edu
🇲🇽🇫🇷🇪🇺 Analytical chemist, specialized in #MassSpec #Proteomics. Leading the Immunopeptidomics Platform at HI-TRON Mainz / DKFZ













