
How DNA secondary structures drive replication fork instability
DNA secondary structures, such as hairpins, cruciforms, triplexes, G-quadruplexes and iMotifs, are common, dynamic features that replication forks rou…
Our @costerlab.bsky.social review is out!
It’s a great read on the impact of DNA secondary structures on eukaryotic replication fork progression - a totally unbiased opinion, of course!
www.sciencedirect.com/science/arti...
@adityasethi.bsky.social @billiedelpino.bsky.social
10.12.2025 11:53 — 👍 16 🔁 8 💬 0 📌 1
![Comparative analysis of the Caulobacter SOS response under mitomycin-C damage. Top left: Schematic summarizing the RNA-sequencing experiment. Caulobacter cells were treated with 0.25 μg/ml mitomycin-C for 20 and 40 min (Created in BioRender). Samples were collected for transcriptomic analysis before (0 min, control), and at 20 and 40 min post damage induction. Top right: Venn diagram representing the genes that meet the listed criteria: 1. Genes induced in wild type cells under MMC damage at 40 min (white circle). 2. Genes induced in ΔlexA in the absence of damage (blue circle). 3. Genes not induced in ΔrecA background under MMC damage at 40 min (gray circle). Genes fulfilling all three criteria are in the gray circle. Number of genes in each category is indicated. Bottom left: Bar graph indicating whether promoters of the shortlisted genes exhibit binding by the LexA protein as assessed from ChIP-seq analysis [10]. Bottom right: LexA ChIP-seq profile for genes belonging to the SOS response. Normalized reads (in rpm) are represented for 500 bp upstream and downstream of the gene CDS.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:5522ztebtekoor5efelihqhb/bafkreigerw7qsbl4br367qyqg3mouvx3va6s7f3pcqugpf2m24adc3wa6y@jpeg)
Comparative analysis of the Caulobacter SOS response under mitomycin-C damage. Top left: Schematic summarizing the RNA-sequencing experiment. Caulobacter cells were treated with 0.25 μg/ml mitomycin-C for 20 and 40 min (Created in BioRender). Samples were collected for transcriptomic analysis before (0 min, control), and at 20 and 40 min post damage induction. Top right: Venn diagram representing the genes that meet the listed criteria: 1. Genes induced in wild type cells under MMC damage at 40 min (white circle). 2. Genes induced in ΔlexA in the absence of damage (blue circle). 3. Genes not induced in ΔrecA background under MMC damage at 40 min (gray circle). Genes fulfilling all three criteria are in the gray circle. Number of genes in each category is indicated. Bottom left: Bar graph indicating whether promoters of the shortlisted genes exhibit binding by the LexA protein as assessed from ChIP-seq analysis [10]. Bottom right: LexA ChIP-seq profile for genes belonging to the SOS response. Normalized reads (in rpm) are represented for 500 bp upstream and downstream of the gene CDS.
The bacterial #SOSresponse unfolds in a defined temporal order, but how does this arise? @adityakamat.bsky.social @anjbadri.bsky.social &co show that intrinsic #promoter strength, modulated by #SigmaFactor usage, governs timing of SOS gene activation in Caulobacter @plosbiology.org 🧪 plos.io/4oAKf0Q
05.12.2025 10:03 — 👍 20 🔁 8 💬 1 📌 0

Congrats to Viri, who successfully defended her PhD thesis yesterday. Well done Dr. Lopez Jauregui! 🎉🎉💯
28.11.2025 15:38 — 👍 3 🔁 0 💬 0 📌 0
Postdoctoral Fellow - H Yardimci Lab
We are recruiting a postdoctoral fellow to study mechanisms of DNA replication stress using single-molecule imaging tools. If you are excited about microscopy, replisome dynamics, and genome stability, we’d love to hear from you.
Application deadline: 24 December
www.crick.ac.uk/careers-stud...
25.11.2025 11:55 — 👍 31 🔁 26 💬 0 📌 0
Thanks Nitin
20.11.2025 23:40 — 👍 0 🔁 0 💬 0 📌 0
Thanks Stephan.
20.11.2025 17:04 — 👍 0 🔁 0 💬 0 📌 0

Genome integrity relies on rapid recycling of DNA Pol III in bacteria | PNAS
DNA replication requires precise coordination between DNA unwinding and DNA synthesis.
In all domains of life, protein–protein interactions at the ...
Happy to share our most recent work www.pnas.org/doi/10.1073/.... Here we describe how efficient DNA replication in E. coli is dependent on an interaction between SSB-ssDNA and the DNA polymerase (Pol III). Thanks to all the co-authors for their hard work!
20.11.2025 16:54 — 👍 38 🔁 17 💬 2 📌 2

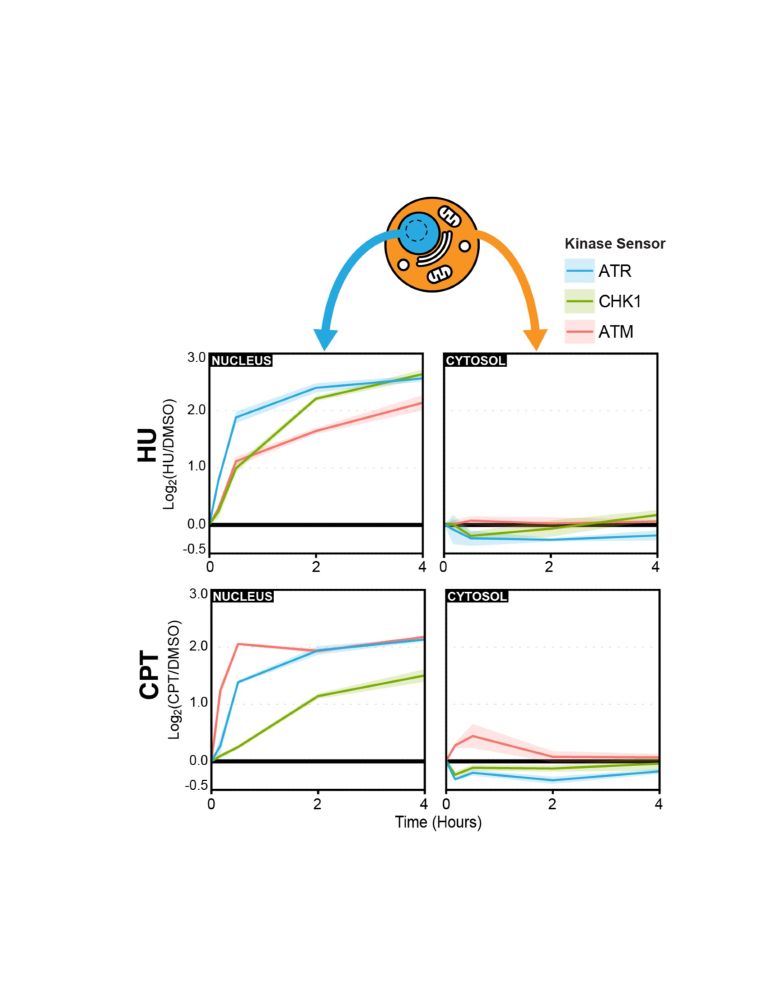
Check out our new biosensor technology to study DDR kinase signaling: ProKAS.
We combine:
-proteomics
-engineered peptide sensors
-a new concept of amino acid barcodes
ProKAS tracks kinase signaling with spatial resolution and produces highly quantitative data.
Just published today: rdcu.be/ePNo0
13.11.2025 18:22 — 👍 21 🔁 8 💬 2 📌 1

Diagram of an E. coli cell showing polysomes pushing sister chromosomes apart. Bottom heatmaps show cell-cycle-dependent
changes in DNA and ribosome density distributions, and constriction formation. The top right corner shows normalized mid cell amounts from these heatmaps (integrated between the dashed horizontal lines).
Our latest work is out in PNAS!🎉
We show that nonequilibrium polysome dynamics help drive sister chromosomes apart. The closing division septum also contributes via steric interactions.👇
www.pnas.org/doi/10.1073/...
#MicroSky #Microbiology #Biophysics #CellBiology
09.11.2025 02:05 — 👍 25 🔁 10 💬 1 📌 1

Modulation of archaeal hypernucleosome structure and stability by Mg2+
DNA-wrapping histone proteins play a central role in chromatin organization, gene expression and regulation in most eukaryotes and archaea. While the …
New article online: Modulation of archaeal hypernucleosome structure and stability by Mg2+
In this work, we dissect the effects of Mg2+ on hypernucleosomes formed by the canonical histones from M. fervidus (HMfA and HMfB) and T. kodakarensis (HTkA and HTkB).
www.sciencedirect.com/science/arti...
06.11.2025 07:53 — 👍 15 🔁 9 💬 1 📌 0

📢 Excited to share that our study has just been published in Nature Communications! We uncover how CRL4-DCAF12 regulates the cellular levels of MCMBP, a chaperone essential for assembling nascent MCM2-7 complexes, to ensure accurate and error-free genome duplication.
28.10.2025 15:04 — 👍 20 🔁 8 💬 4 📌 0
PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
A new, nerdy paper. We figured out (some) of the rules underlying cell-permeability of probes and designed ligands that light up, grab, and move proteins around. Awesome @hhmijanelia.bsky.social x @uwmadison.bsky.social x @stjuderesearch.bsky.social collaboration! www.pnas.org/doi/10.1073/...
27.10.2025 21:43 — 👍 87 🔁 30 💬 0 📌 1
Hello! The Jacobs-Wagner lab is finally on Bluesky. We’re excited to celebrate science (and scientists!), share our research, and interact with the community. We look forward to connecting with friends and colleagues across the world.
19.10.2025 20:06 — 👍 45 🔁 13 💬 2 📌 0
This was a fun collaboration between Ophélie & the @seegerlab.bsky.social using sybody libraries to target the SMC complex in living bacteria. With a suprising finding: the 14 isolated sybodies bind to the middle of the SMC coiled coil rather than the more conserved ATPase heads.
20.10.2025 08:15 — 👍 7 🔁 5 💬 0 📌 0
Now in @mdpiopenaccess.bsky.social Bacillus subtilis DinG/XPD-like paralogues, DinG and YpvA, have been implicated in overcoming replication stress. DinG possesses a DEDD exonuclease and DNA helicase domains, whereas YpvA lacks the DEDD exonuclease domain
www.mdpi.com/1422-0067/26...
04.10.2025 10:51 — 👍 2 🔁 2 💬 0 📌 0

Postdoctoral Fellow - Costa Lab
Salary for this Role: From £45,500 with benefits, subject to skills and experience. Job Title: Postdoctoral Fellow - Costa Lab Reports to: Alessandro Costa Closing Date: 03/Aug/2025 23.59 GMT Job Desc...
Less than two weeks left to apply!
Postdoc positions in my lab to study
1. initiation of DNA replication.
2. chromatin replication/epigenetic inheritance.
Great for biochemists, biophysicists & structural biologists.
Deadline 3 August 2025.
crick.wd3.myworkdayjobs.com/External/job...
21.07.2025 14:57 — 👍 25 🔁 31 💬 0 📌 0
Head of Genome Biology Dept @EMBL,
Scientist, Principal investigator, Professor
Exploring genome regulation during development,
and everything to do with enhancers.
3D genome, chromatin topology,
cell_fate, embryonic Development,
Single Cell genomics
Studying archaeal 3D genomes at Ritsumeikan University, Japan
https://scholar.google.co.jp/citations?user=Q34_N9wAAAAJ&hl=ja#
(he/him) Postdoc | Genome Replication Lab at Institute of Cancer Research London https://costerlab.com/ | 🏳️🌈
Postdoctoral scientist | JIC, UK
PhD | de Duve institute, Belgium
Passionate about killing E. coli and watching them die under the🔬
Our group at the LMB in Cambridge is dedicated to uncovering structures and mechanisms of the replisome.
https://www2.mrc-lmb.cam.ac.uk/group-leaders/t-to-z/joseph-yeeles/
Executive Editor/Team Leader Open Access Science Journals Sage Publishing
Opinions = mine
http://linkedin.com/in/jlovick-editor
#oncology #cancerresearch #medicine #biology #cardiology #neurology #microbiology #publichealth #healthcare #technology
Postdoc at NIH/NCI, PHD at Hopkins
CNRS researcher in the pneumococcal competence and transformation team in Toulouse.
Using molecular genetics and cell biology to study pneumococcal competence and transformation.
Postdoc in the Cell Biology and Virology of Archaea Unit @pasteur.fr
Archaea is the coolest domain of life! 🔬🦠
PPU PhD @pasteur.fr, Mart Krupovic
MSc Microbiology @helsinki.fi, Mikael Skurnik
BSc Biomedicine @UB.edu
In our lab, hardcore biochemistry meets physics. And coffee. We are investigating DNA replication through single molecule loupe @ Technical University of Munich and MPI of Biochemistry
Associate Professor. NC State. Microbial Genomics, Evolution, Bioinformatics.
Postdoc at Vanderbilt University studying how bacteria mutate, evolve, and repair their DNA. PhD in Genetics and Molecular Biology from The University of North Carolina. Biotecnólogo por la Universidad de Oviedo. Asturiano en Tennessee.
Molecular microbiologist intrigued by all things bacteria.🧫🦠🌱🪲🧬🧪Research Scientist. Current focus: genetics, ecology, and evolution of bacterial plant pathogens. Opinions expressed are solely my own.
ORCID: 0000-0002-2723-8217
Circadian Sleep Disorders Advocate. Raising awareness for Non24 in sighted people. #DSPD mom of a #Non24 teen. Former board member at https://csd-n.org.
🦠🧪🖥🧬microbial ecology & evolution | agricultural microbiomes | food security | metabolism | 🐝🌺 host-microbe interactions | functional -omics | bioinformatics | salsa | philosophy | equality | UNAM | mSystems / mBio
polyimmigrant! 🇲🇽🇸🇪🇦🇺🇨🇭
Microbiologist, Research, Molecular genetics, DNA, Genome dynamics
#Evolution, #Microbiology, and #Phages. Associate Professor, University of Canterbury, Christchurch NZ. Adaptable Phage Solutions Co-Science Lead.Views my own. |She| Her|.
hendricksonlab.co.nz
We investigate the process of chromosomal DNA replication and its impact on genome function and stability using molecular genetics and strand-specific genomics
Bertolin lab | Understanding genomic stability and human disease through a DNA replication lens | Wellcome-funded Group Leader, School of Life Sciences, Dundee, Scotland | Former Diffley lab postdoc, Francis Crick Institute


![Comparative analysis of the Caulobacter SOS response under mitomycin-C damage. Top left: Schematic summarizing the RNA-sequencing experiment. Caulobacter cells were treated with 0.25 μg/ml mitomycin-C for 20 and 40 min (Created in BioRender). Samples were collected for transcriptomic analysis before (0 min, control), and at 20 and 40 min post damage induction. Top right: Venn diagram representing the genes that meet the listed criteria: 1. Genes induced in wild type cells under MMC damage at 40 min (white circle). 2. Genes induced in ΔlexA in the absence of damage (blue circle). 3. Genes not induced in ΔrecA background under MMC damage at 40 min (gray circle). Genes fulfilling all three criteria are in the gray circle. Number of genes in each category is indicated. Bottom left: Bar graph indicating whether promoters of the shortlisted genes exhibit binding by the LexA protein as assessed from ChIP-seq analysis [10]. Bottom right: LexA ChIP-seq profile for genes belonging to the SOS response. Normalized reads (in rpm) are represented for 500 bp upstream and downstream of the gene CDS.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:5522ztebtekoor5efelihqhb/bafkreigerw7qsbl4br367qyqg3mouvx3va6s7f3pcqugpf2m24adc3wa6y@jpeg)


















