Let me know if you’d like me to clarify anything. I’m happy to talk!
25.05.2025 20:54 — 👍 0 🔁 0 💬 0 📌 0
Me too 🤪 It is really exciting to be submitting! We definitely learned a lot along the way
10.05.2025 05:27 — 👍 2 🔁 0 💬 0 📌 0


Reinforcement learning with experimental feedback (RLXF) shifts protein language models so that they generate sequences with improved properties
@nathanielblalock.bsky.social @philromero.bsky.social
www.biorxiv.org/content/10.1...
10.05.2025 01:46 — 👍 38 🔁 7 💬 1 📌 0
Thank you for sharing our work @kevinkaichuang.bsky.social! It means a lot
10.05.2025 02:05 — 👍 1 🔁 0 💬 1 📌 0
Thank you for posting about our preprint!
08.05.2025 18:03 — 👍 1 🔁 0 💬 0 📌 0
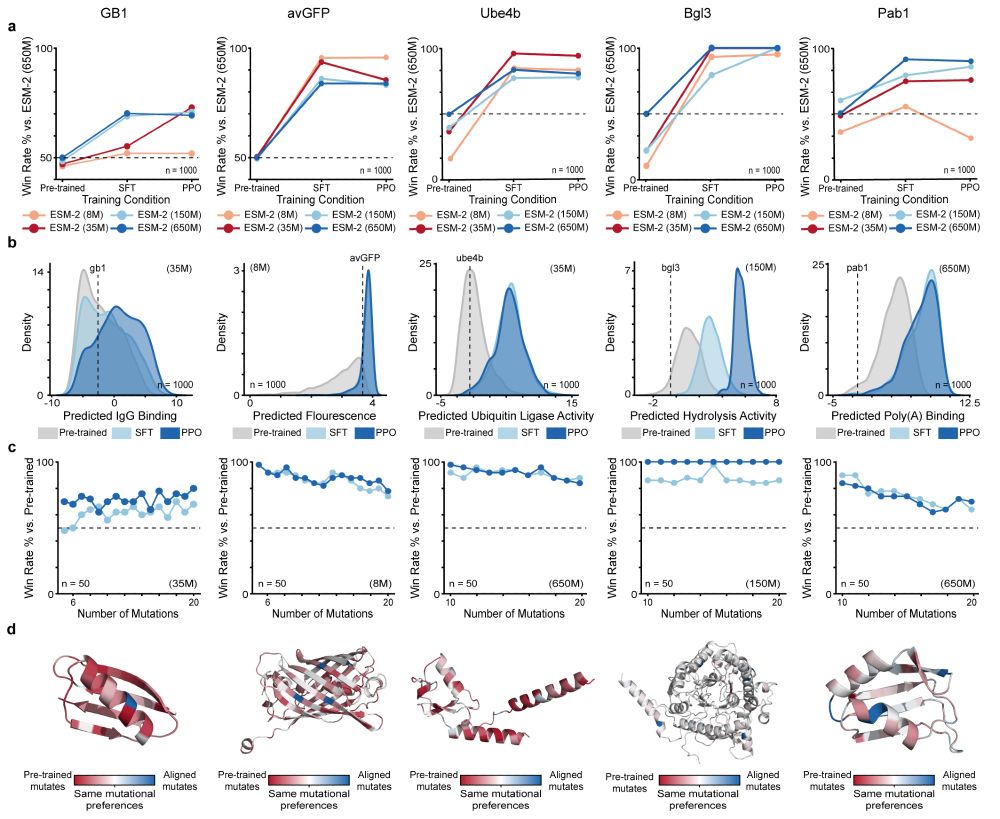
We apply RLXF across five diverse protein classes to demonstrate its generalizability and effectiveness at generating optimized sequences by learning functional constraints beyond those captured during pre-training
08.05.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0

Experimental validation reveals the RLXF-aligned model generates a higher fraction of functional sequences, a greater number of sequences more fluorescent than CreiLOV, and the brightest oxygen-independent fluorescent protein variant reported to date
08.05.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0

We align ESM-2 to experimental fluorescence data from the CreiLOV flavin-binding fluorescent protein. The aligned model learns to prioritize mutations that enhance fluorescence, many of which are missed by the base model
08.05.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0

RLXF follows a two-phase strategy inspired by RLHF. Supervised Fine-Tuning initializes the model in the right region of sequence space. Proximal Policy Optimization directly aligns sequence generation with feedback from a reward function like a sequence-function predictor
08.05.2025 18:02 — 👍 0 🔁 1 💬 1 📌 1
Pre-trained pLMs generate highly diverse sequences mirroring statistical patterns from natural proteins. But here's the challenge: they lack an explicit understanding of function, often failing to generate proteins with enhanced or non-natural activities. RLXF bridges this gap!
08.05.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0

We are excited in the @philromero.bsky.social lab to share our new preprint introducing RLXF for the functional alignment of protein language models (pLMs) with experimentally derived notions of biomolecular function!
08.05.2025 18:02 — 👍 3 🔁 0 💬 1 📌 1
Post the amazing science things you have done with federal funding.
28.01.2025 20:51 — 👍 1558 🔁 603 💬 172 📌 319
It was a pleasure meeting you! Y'all are doing super interesting and relevant work. It will be cool to see how we can continue to interact and maybe collaborate in the future!
20.12.2024 20:50 — 👍 2 🔁 0 💬 0 📌 0
My 1st NeurIPS was a wonderful experience - incredible to see so much research in protein design and reinforcement learning. Here are my favorite papers (and favorite places I got food in Vancouver 😋):
20.12.2024 16:42 — 👍 3 🔁 0 💬 1 📌 0
Hey Kevin, could I be added? This is really helpful for joining Bluesky! Thank you for doing it
17.12.2024 18:38 — 👍 1 🔁 0 💬 0 📌 0
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
03.12.2024 03:27 — 👍 147 🔁 60 💬 16 📌 6
Bringing unparalleled #accuracy and precision to #drugdiscovery and design, using #quantumphysics to develop life-changing treatments for major diseases.
Visit our website: https://www.qubit-pharmaceuticals.com/
Learn, explore, lead, and change. Love most things at the subcellular level. Manager in CAR-T Operations, avid reader, practical joker.
MSCA fellow at @crg.eu w/M. Dias and @jonnyfrazer.bsky.social
> Biological Physics | Proteins | Comp Bio | ML
https://scholar.google.com/citations?user=n55NtEsAAAAJ&hl=en
PhD student within the Functional Genomics group (Franke lab), University Medical Centre Groningen. Interested in gene networks, sequence-based models and non-coding somatic mutations
UofArizona #Aggie > UofMaryland PhD > NIH/NCI PostDoc > Biodefense, Pathogen #Genomics, #Bioinformatics, Data Authenticity, #BioSecurity. ++PC gaming, DnD, EDM, punk, goth, backcountry backpacking, porsche
🎲♟️👾🎮 🎸 🧪 🧬 🖥 🦠🚀🏎️
he/him. On Steam: @bioinformer
PhD at EPFL | Biophysics and Machine Learning
Thesis student with @larsplus.bsky.social and @rmartinezcorral.bsky.social at @crg.eu Prev: IISER Pune
(he/him)
PhD student @sangerinstitute
From Pipette to Python | Drug & Protein Design | Structural Biology
Postdoc // gene regulation, genomics, synbio, stem cells, c. elegans // with @eddaschulz @molgen.mpg.de previously @n_rajewsky @mdc-bimsb //
researching biological AI-applications
https://moritzgls.github.io/
Senior Systems Engineer / Devops Engineer / Science Nerd in Vancouver, Canada #SlavaUkraini #StandWithUkraine🇺🇦
AI scientist, roboticist, farmer, and political economist. Governments structure markets. IP is theft. @phytomech.com is my alt.
https://advanced-eschatonics.com
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
Industrial biotech and enzymes @ Novonesis 🦠🔬. Solarpunk and cocktails from here on out 🎋🍹.
scientist at UC Berkeley inventing advanced genomic technologies
lover of molecules, user of computers
https://scholar.google.com/citations?user=63ZRebIAAAAJ&hl=en
Data, biotech, engineering, biology, aws
Science, housing advocacy, transit, Lego. 🧬🤖🌱 Engineering Biology Platform Lead, Global Institute for Food Security
https://scholar.google.ca/citations?user=dHPm9s0AAAAJ&hl=en















