Tres miembros de la RSEQ han conseguido una ERC... – RSEQ
Los miembros de la RSEQ: Carla Casadevall, Nicoletta Liguori y Carlos Moreno Yruela consiguen una ERC Starting Grant en la convocatoria 2025.
🎉 ¡Orgullo RSEQ! Nuestros socios @casadevallcarla.bsky.social,
@nikinikki87.bsky.social y @carlosmyruela.bsky.social han sido galardonados con una prestigiosa #ERCStartingGrant 2025🔬 Enhorabuena!!!!!
📖 rseq.org/tres-miembro...
@urv.cat
@lcbm-epfl.bsky.social
@iciq.org @icfo.eu
10.09.2025 13:44 — 👍 7 🔁 5 💬 0 📌 0
I am really excited that #CHEMTUBIO will be funded through an @erc.europa.eu Starting Grant!
This funding will kickstart my lab next year and help us unveil the mechanisms and therapeutic implications of erasers of microtubule modifications.
Stay tuned!
05.09.2025 11:24 — 👍 7 🔁 0 💬 0 📌 0

Six #BISTcommunity early-career researchers have secured prestigious ERC grants 👏
These awards highlight the ability of BIST Community centres to attract and nurture outstanding young international talent.
@crg.eu @icfo.eu @iciq.org @ibecbarcelona.eu
👉http://bit.ly/486ENOq
04.09.2025 15:08 — 👍 9 🔁 2 💬 0 📌 1
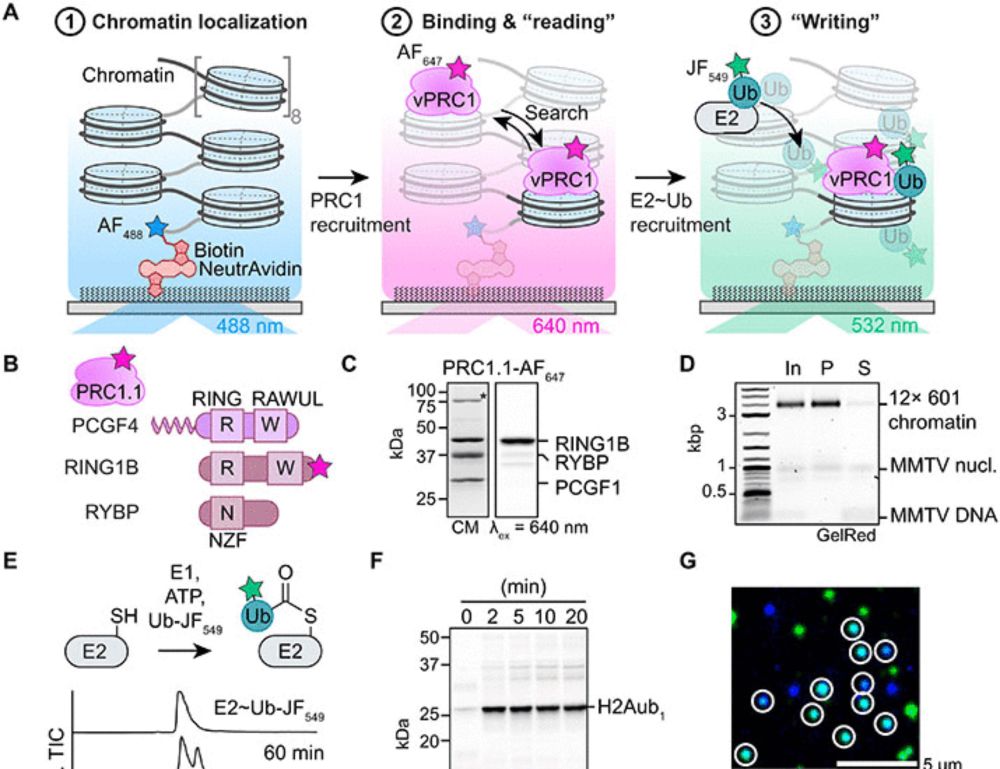
Single-molecule analysis reveals the mechanism of chromatin ubiquitylation by variant PRC1 complexes
Single-molecule experiments show that active conformation formation controls chromatin ubiquitylation kinetics by variant PRC1.
If you always wanted to know the kinetics of how the Polycomb enzymes modify chromatin, we are thrilled to share our work!!! 🎉📄🧬🔬
Thank you, @beatfierz.bsky.social for all the guidance as well as the @lcbm-epfl.bsky.social group members for all the support!
doi.org/10.1126/scia...
22.05.2025 11:00 — 👍 25 🔁 5 💬 1 📌 0
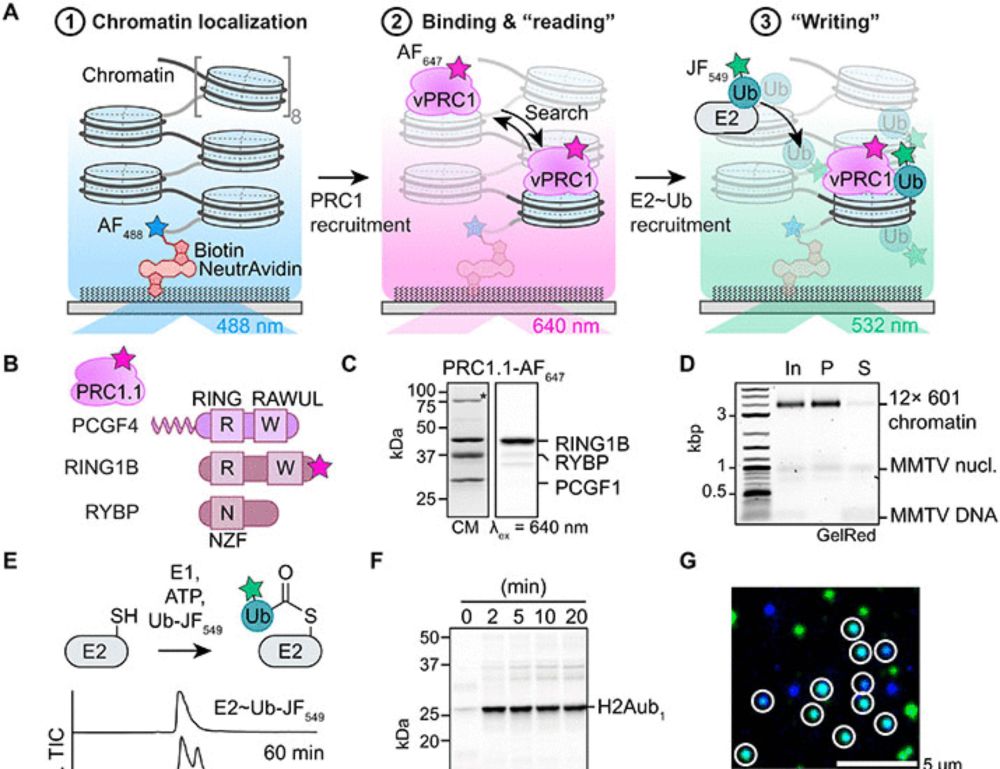
Single-molecule analysis reveals the mechanism of chromatin ubiquitylation by variant PRC1 complexes
Single-molecule experiments show that active conformation formation controls chromatin ubiquitylation kinetics by variant PRC1.
Check out our new study of chromatin ubiquitylation. @alexateslenko.bsky.social visualized directly at single molecule scale how vPRC1 ubiquitylates neighboring nucleosomes. These results suggest potential mechanism on how H2Aub domains are established. 👇👇👇
www.science.org/doi/10.1126/...
22.05.2025 10:47 — 👍 31 🔁 8 💬 0 📌 2

Lausanne
Explore Pint of Science Switzerland events in Lausanne
The program for Pint of Science Lausanne 🧪 is ready!
pintofscience.ch/events/lausa...
This year we have again three venues covering topics from aging to robotic surgery or sustainable agriculture
Come join us on the 19th, 20th and 21st of May 🥳
#PintCH #scicomm @pintofscience.ch
24.04.2025 08:57 — 👍 1 🔁 0 💬 0 📌 1
What started as the M.Sc. projects of Michaela Benová and Athanasios Tsiris, seeking potent and selective inhibition of #HDAC11, turned into a great collaboration between the labs of @christianaolsen.bsky.social and Christian Heinis led by @danieladankova.bsky.social.
Big congratulations, Daniela!
19.02.2025 11:54 — 👍 3 🔁 0 💬 1 📌 0

!THREE OPPORTUNITIES TO JOIN US!
We are looking for 3 (!!!) PhD students interested in:
How Metabolism affects Genome Regulation.
Join us at @carrerasijc.bsky.social as Marie Sklodowska Curie network fellow.
More info:
2 x experimental: www.nuclear-dn.eu
1 x computational: www.hubmol.eu
12.02.2025 15:34 — 👍 8 🔁 11 💬 0 📌 1
🚨 Excited to share our work @elife.bsky.social doi.org/10.7554/eLife.98827.2 on targeting Hepatitis B virus #HBV 🦠 capsid aggregation by designed molecules! #DrugDiscovery #ChemBio
Thanks V Khayenko C Makbul @clemensschulte.bsky.social & @boettchercryoem.bsky.social for stunning #CryoEM visuals!
28.01.2025 19:07 — 👍 16 🔁 3 💬 0 📌 1
Thanks, Christian!
04.02.2025 20:24 — 👍 0 🔁 0 💬 0 📌 0
@naturecomms.bsky.social
04.02.2025 13:46 — 👍 1 🔁 0 💬 0 📌 0
Thank you as well to @snsf-ch.bsky.social and IRFD for funding my postdoc at EPFL, to the @swisschemistry.bsky.social foundation and La Caixa Foundation for supporting Polina and Esther, and other funding from @snsf-ch.bsky.social, EPFL and the Dubochet Center for Imaging.
(8/8)
04.02.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0

This would not have been possible without the excellent (and very efficient!) contributions of Babatunde Ekundayo and Polina Foteva (who just defended her MSc thesis 🥳), of Dongchun Ni, Esther Calviño and Henning Stahlberg, or without the truly supportive guidance of @beatfierz.bsky.social.
(7/8)
04.02.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0

The contacts of the catalytic domain with DNA, on the other hand, are responsible for maintaining #H3K36ac selectivity.
Mutating a key interacting loop helps release the enzyme from the nucleosome and become much more active on #H3K18ac substrates 🤯.
(6/8)
04.02.2025 13:13 — 👍 2 🔁 0 💬 1 📌 0

The multivalent interactions of the nucleosome-binding domain are important for both activities, especially to find #chromatin substrates in an environment full of nucleic acids.
Interestingly, this domain is only present in insects and most vertebrates.
(5/8)
04.02.2025 13:13 — 👍 2 🔁 0 💬 1 📌 0

By comparing structures trapped at each substrate position we could rationalize its activity and intrinsic selectivity 🔍.
#SIRT7 binding is optimal for targeting #H3K36ac, and it can only access #H3K18ac through partial disengagement from the nucleosome and DNA bending.
(4/8)
04.02.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0

Babatunde Ekundayo generated beautiful high-resolution structures, where we identified:
- A unique N-terminal nucleosome-binding domain, which places the #SIRT7 catalytic domain by the H3 tail exit site
- Extensive contacts with the two DNA gyres!
(3/8)
04.02.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0

We produced nucleosomes with a thiourea mechanism-based handle (MTU) at the H3K18 and H3K36 preferred substrate positions, through histone semi-synthesis and reconstitution in vitro.
This allowed us to stabilize specific enzyme:substrate complexes efficiently for #cryoEM.
(2/8)
04.02.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0
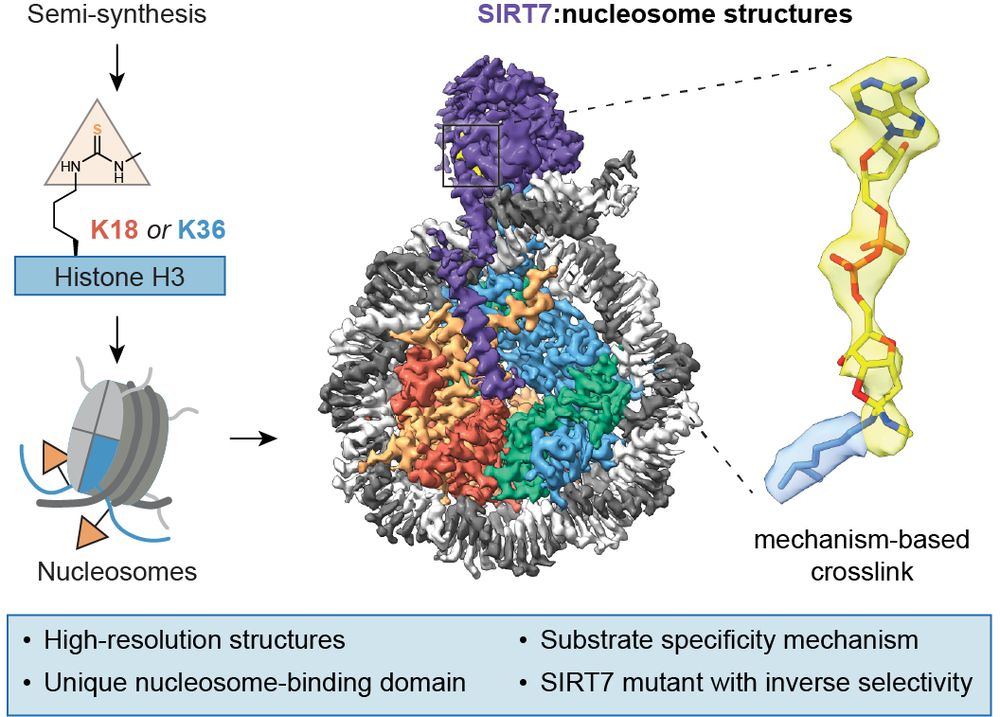
#SIRT7 is a histone deacetylase with highly specific activity on #chromatin substrates.
We just published mechanism-based #cryoEM structures of #SIRT7 on nucleosomes to understand its activity 👇
www.nature.com/articles/s41...
(1/8) #ChemBio #ChemSky
04.02.2025 13:13 — 👍 43 🔁 17 💬 1 📌 1
Hi Dorien, I would love to be added to this started pack!
25.01.2025 11:36 — 👍 1 🔁 0 💬 1 📌 0

High-resolution structural imaging using customised magnetic beads.
Cryogenic Electron Microscopy: MagIC beads for scarce macromolecules. #CryoEm
https://elifesciences.org/articles/105335?utm_source=bluesky&utm_medium=social&utm_campaign=organic_insights
25.01.2025 09:21 — 👍 9 🔁 4 💬 0 📌 0
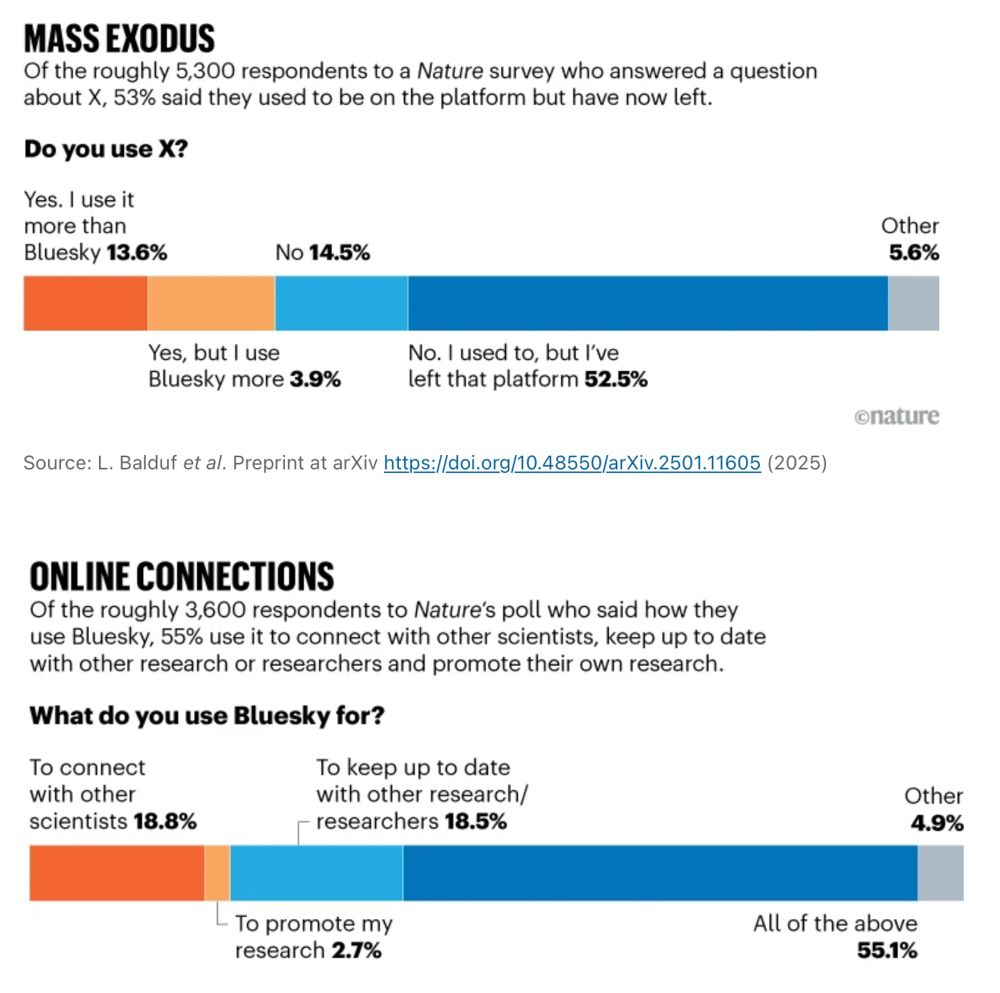
Welcome immigrants from X interested in science to the friendlier skies of @bsky.app, as documented by a new @nature.com survey (but you already knew that 😉)
"Bluesky is much better for science. There is much less toxicity, misinformation, and distractions."
www.nature.com/articles/d41...
24.01.2025 15:22 — 👍 15548 🔁 2399 💬 247 📌 119
MagIC-Cryo-EM: Structural determination on magnetic beads for scarce macromolecules in heterogeneous samples
Here is our most recent reviewed preprint on MagIC-cryo-EM invented by @yarimura.bsky.social
elifesciences.org/reviewed-pre...
Please note that this is not the final version, or VOR. A revised version (www.biorxiv.org/content/10.1...) is currently being reviewed at eLife.
22.01.2025 18:31 — 👍 3 🔁 1 💬 1 📌 0
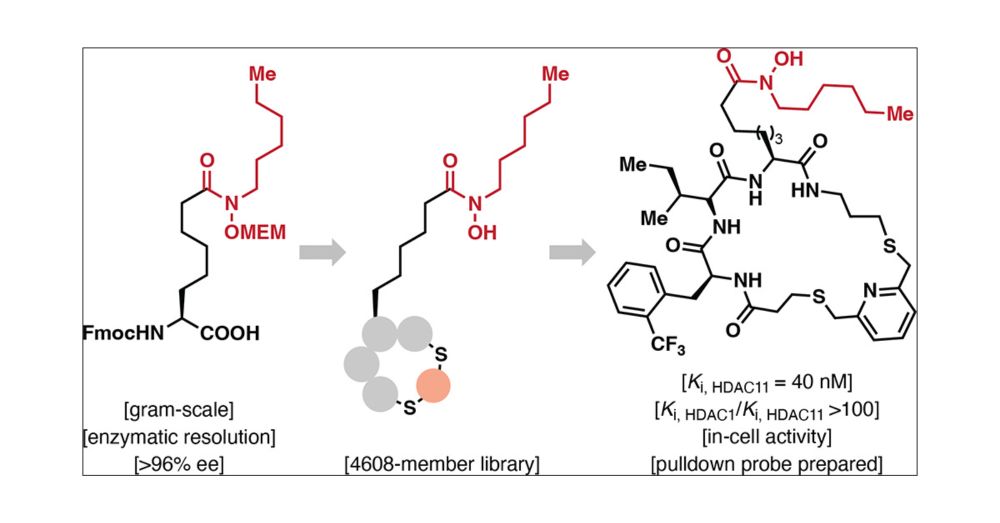
Happy to share the latest preprint from my PhD studies on the discovery of de novo macrocyclic inhibitors of HDAC11. This was a great collaboration between @christianaolsen.bsky.social lab (@carlosmyruela.bsky.social, @tnhansen.bsky.social) and Christian Heinis lab (@alexanderln.bsky.social).
13.12.2024 07:10 — 👍 11 🔁 3 💬 1 📌 1
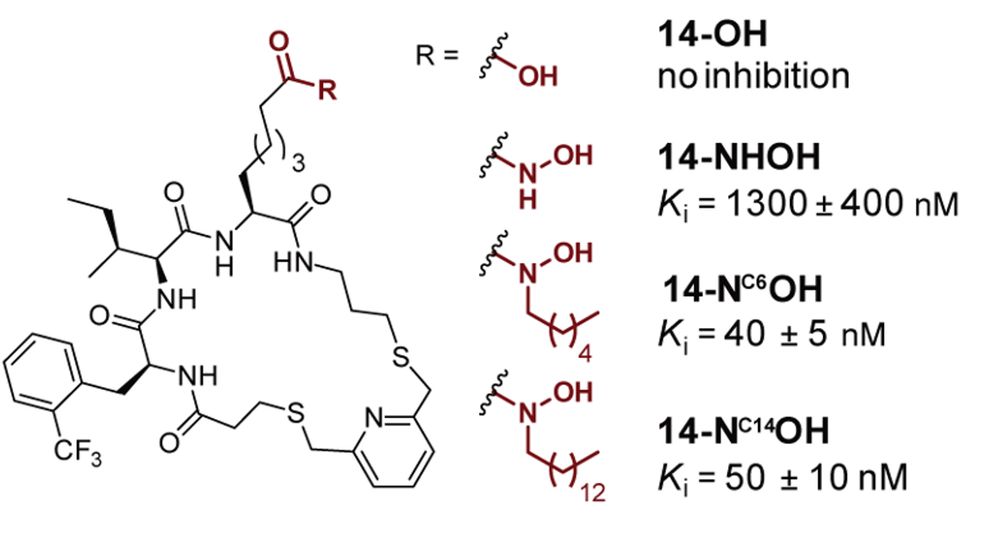
New preprint @chemrxiv.bsky.social on discovery of macrocyclic inhibitors of HDAC11 with the Christian Heinis #EPFL
Main contributors @danieladankova.bsky.social, @alexanderln.bsky.social, @carlosmyruela.bsky.social
🙏 @ERC_Research, @novonordiskfond, @DFF_raad for 💶
chemrxiv.org/engage/chemr...
10.12.2024 14:27 — 👍 18 🔁 6 💬 0 📌 3
Chemoproteomics @ Athens, Greece
#ChemBio
#chemoproteomics
https://orcid.org/0000-0002-4782-4029
Postdoc @UoD @CeTPD with Alessio Ciulli
Structural & chemical biology in Targeted Protein Degradation
La RSEQ es una sociedad, declarada de utilidad pública, que tiene por objeto promover, desarrollar y divulgar la disciplina de la Química, tanto en su aspecto de ciencia pura como en el de sus aplicaciones, en todo el ámbito del estado español.
De novo evolved proteins. @bornberglab.bsky.social. he/him.
bornberglab.org/people/eicholt/
LMU Munich
Chemical Biology of Protein Post-Translational Modifications
Chemoproteomics, Click Chemistry, Mass Spectrometry
Postdoctoral Research Fellow at Marc Vendrell's Lab @Dynafluors @EdinburghUni. PhD at @STHenriquesLab & @theCraikGroup. Dad.
Chemical Biology; Drug Delivery; Antibody-Drug Conjugates; Bioactive Peptides
Group leader @EMBL Heidelberg | Archaea | Chromatin | Cryo-EM and cryo-ET | Evolution | Structure
Passionnate about microscopy and sharing science @PintofScience.ch
🇲🇫 Postdoc at the University of Lausanne 🇨🇭
Plant-Microbiota-Pathogen interactions | Bacterial Ecology | Microfluidics | Single-cell imaging | Systems biology
https://julienluneau.com
Researcher in Epigenetics, CRISPRing around. MSCA Postdoctoral Fellow at EMBL Rome (Hackett lab). Before: Max Planck for Molecular Genetics (Ph.D.), Exeter Uni, Pasteur Institute, Sapienza Uni - Pessimismo dell' intelligenza/Ottimismo della volontà.
Laboratory of Biophysical Chemistry of Macromolecules (LCBM) headed by @beatfierz.bsky.social at EPFL | Run by LCBM members
For more information and registration visit: www.divide-conference.com
SNSF postdoctoral fellow EPFL
EPFL Postdoc Association advisor & event mastermind 🎉
Passionate chemist harnessing light to unlock molecular secrets ✨🔬
Nature lover, peace seeker, and adventure enthusiast 🌿☀️
An international journal at the heart of open access for the global chemistry community.
Published by @rsc.org 🌐 Website: rsc.li/rscadvances
Professor at University Frankfurt
Scientist, Biomolecular Chemist, Protein Engineer
Biochemistry and Bioengineering of Megasynthases
Biochemist 🧪| Empowering Life Scientists and Science Communicators to Achieve Their Career Goals | UCLA PhD + NIH Alum | Pharma R&D @GSK + @AstraZeneca 💊 | #SciComm Director ✍️| Career Coach | Star Trek | Sherlock Holmes | 🎹 | https://larrymillerphd.com
Chemistry at UCLA, she/her
SlettenGroup.chem.ucla.edu
Postdoc in chromatin chemical biology at Princeton University.
Empowering scientists with tools that reveal crucial insights not available before, at the molecular and cellular level | Dynamic single-molecule & Cell Avidity