Scientists have seen Asgard archaea crawling for the first time. When it comes to the origin of eukaryotes, this is like seeing a feathered dinosaur in the wild. (Video courtesy of Philipp Ralder)
18.02.2026 20:09 — 👍 467 🔁 136 💬 9 📌 18
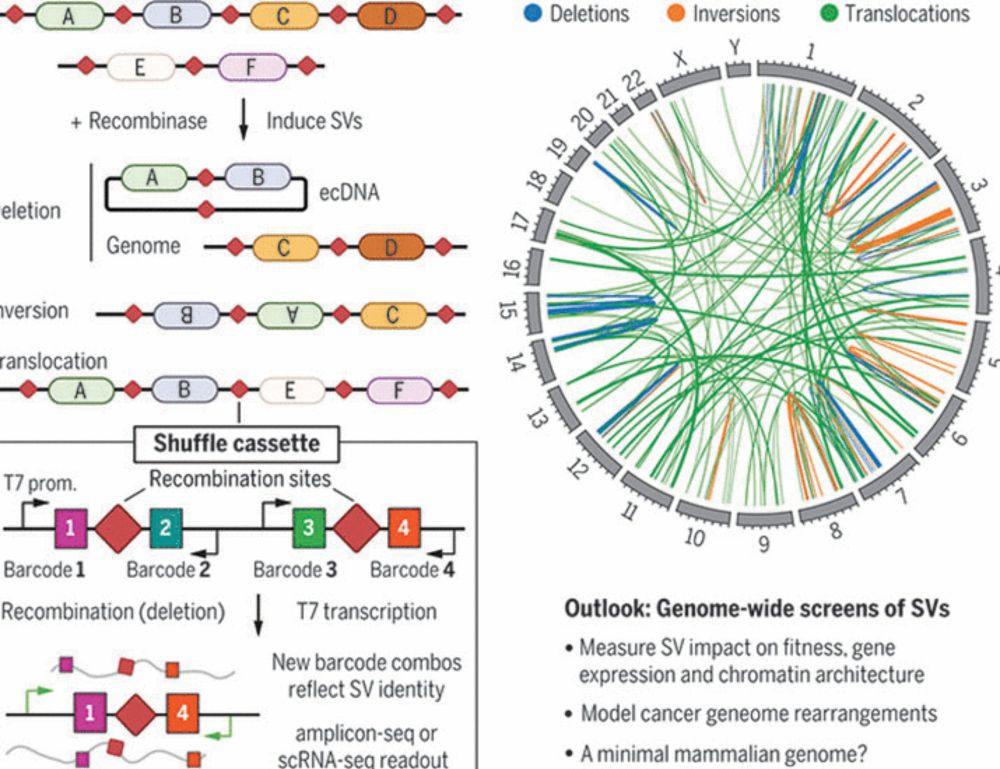
Very happy to share our latest led by the relentless @shawnfayer.bsky.social! We developed multiplexed assays to measure variant effects across diverse genetic and cell contexts in stem cell and differentiated cells. 🧵 👇
22.11.2025 17:08 — 👍 4 🔁 1 💬 0 📌 0

Karel Svoboda and Jay Shendure elected to National Academy of Medicine
The honor recognizes leaders who have demonstrated outstanding achievement and made lasting contributions to the advancement of the medical sciences, health care, and public health
Congratulations to Karel Svoboda and @jshendure.bsky.social on their election to the National Academy of Medicine (@nam.edu)! Election to the Academy is one of the highest honors in health and medicine. #NAMmtg
More on their journeys to this achievement: alleninstitute.org/news/karel-s...
20.10.2025 19:15 — 👍 51 🔁 10 💬 1 📌 0


Super excited about first Shendure/Baker Lab collaboration & preprint on a multiplex sequencing-based strategy for screening de novo proteome editors in mammalian cells. Kudos to the brilliant Chase Suiter (not here) & @greenahn.bsky.social on the work! Preprint here:
www.biorxiv.org/content/10.1...
14.10.2025 18:44 — 👍 97 🔁 33 💬 0 📌 0

New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...
26.09.2025 18:06 — 👍 54 🔁 20 💬 3 📌 2

It’s been a tough few weeks. My 10yo daughter was diagnosed with a very rare, aggressive cancer called interdigitating dendritic cell sarcoma (IDCS). I’m reaching out to identify clinicians/patients who have encountered pediatric IDCS or other (non-LCH) dendritic or histiocytic sarcomas cases.
08.02.2025 21:21 — 👍 1010 🔁 849 💬 83 📌 32
Thanks GSA for blogging about our work!
10.05.2024 20:09 — 👍 1 🔁 0 💬 0 📌 0
Washington Research Foundation announces its largest ever cohort of WRF Postdoctoral Fellows for 202...
Very grateful to be amongst this amazing cohort of scientists! Many thanks to @wrfseattle for the opportunity to learn and grow in this diverse program. Finally, I can't thank my mentors @jshendure.bsky.social and @lea_starita at UW Genome Sciences enough for their support!
14.12.2023 22:17 — 👍 1 🔁 0 💬 0 📌 0
If you're interested in hearing more about this work, I will be speaking at the #2023MasterBrewers Meeting in October! Please come by!
23.09.2023 18:18 — 👍 3 🔁 1 💬 0 📌 0
Finally, I can't thank my mentors @edwardmarcotte.bsky.social and @maitreya.bsky.social enough for all their support and encouragement without which none of this would have been possible!
23.09.2023 18:18 — 👍 2 🔁 0 💬 1 📌 0

Always grateful to Chip, Dusan and Thorin @LiveOakBrewing (Twitter) for enthusiastically supporting us even through tough pandemic times and showing me that interesting scientific questions aren't restricted to the lab.
23.09.2023 18:17 — 👍 2 🔁 0 💬 1 📌 0
A huge thank you to my fellow authors, @reneegeck.bsky.social, Joe Armstrong, Barbara Dunn, Daniel Boutz, Anna Battenhouse, @MarioLeutert (Twitter),
@vyqtdang (Twitter), @pyjiang (Twitter), Dusan Kwiatkowski, Thorin Peiser, and Chip McElroy for their help and assistance!
23.09.2023 18:16 — 👍 2 🔁 0 💬 1 📌 0
We're really excited to share this resource with the community, and thanks to @MarioLeutert (Twitter), we have this website for exploring the data! brewing-yeast-proteomics.ccbb.utexas.edu
23.09.2023 18:13 — 👍 1 🔁 0 💬 1 📌 0


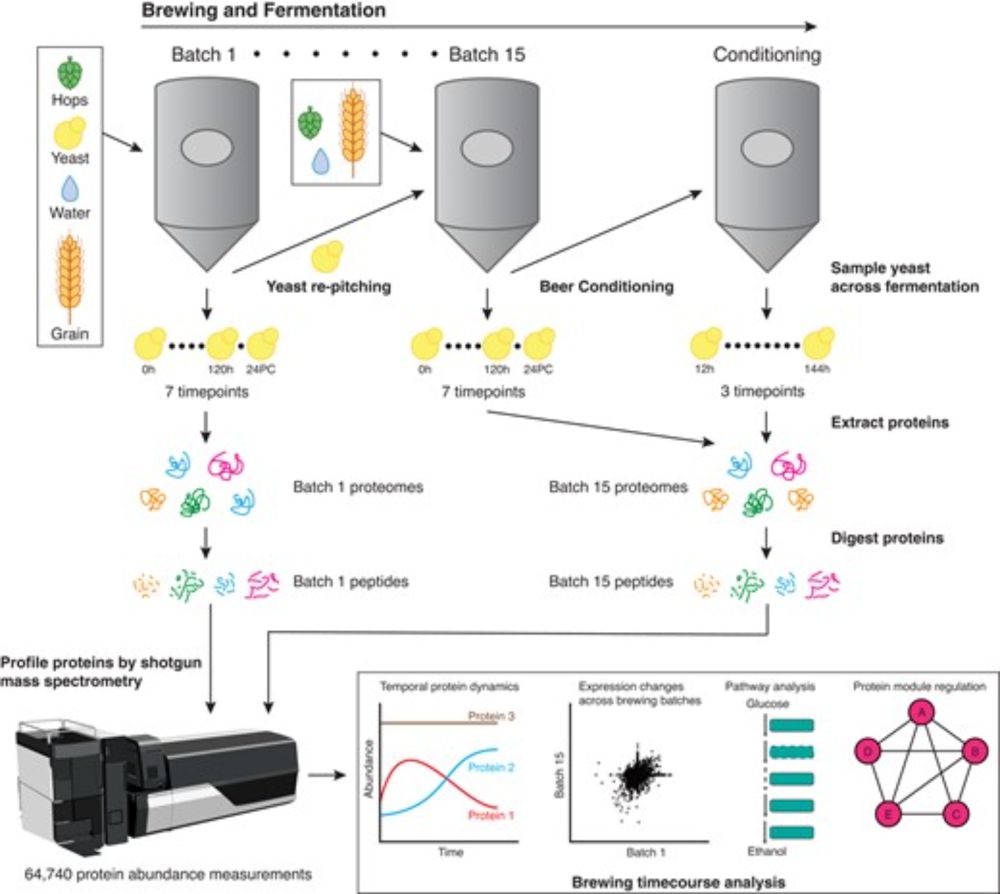
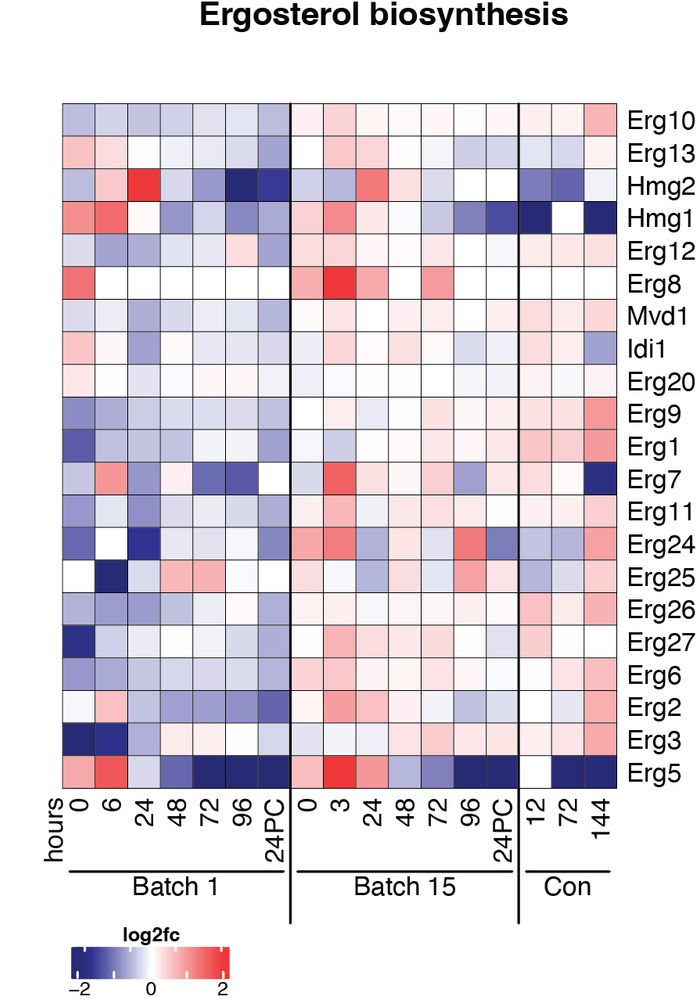
So what did we learn from making over 64,000 protein abundance measurements across 17 different timepoints? -We now have a global view of the temporal protein changes in yeast across brewing including when and which processes change most between timepoints.
23.09.2023 18:10 — 👍 2 🔁 0 💬 1 📌 0

Sample collection was particularly fun because it required sampling directly from fermentation tanks!
23.09.2023 18:09 — 👍 2 🔁 0 💬 1 📌 0

Our work directly profiles the actual brewing yeast populations from fermentation tanks to understand how yeast proteomes temporally (and globally) change over fermentation. We used proteomics to profile brewing yeast both across fermentation cycles and serial repitching.
23.09.2023 18:08 — 👍 2 🔁 0 💬 1 📌 0
The majority of previous fermentation yeast omics studies characterize the domestication history, genetic differences across strains, and understand how proteins and metabolites in beer influence flavor.
23.09.2023 18:07 — 👍 2 🔁 0 💬 1 📌 0
It was purely a labor-of-love driven by curiosity and conversations with biochemist and founder Chip McElroy from @LiveOakBrewing (Twitter) wanting to know what molecular changes yeast cells go through during fermentation and brewing.
23.09.2023 18:06 — 👍 3 🔁 0 💬 1 📌 0
Finally on BlueSky! Thrilled to share our work profiling Ale yeast protein dynamics across fermentation and serial repitching! biorxiv.org/content/10.1...
@maitreya.bsky.social @edwardmarcotte.bsky.social
@LiveOakBrewing #BeerYeast #SystemsBiology #Proteomics Sneek Peak🧵 below-
23.09.2023 18:06 — 👍 14 🔁 3 💬 1 📌 1
Super fun project with @riddhimankg (twitter), @edwardmarcotte.bsky.social, @LiveOakBrewing (twitter), and friends! Systematic Profiling of Ale Yeast Protein Dynamics across Fermentation and Repitching
www.biorxiv.org/content/10.1...
22.09.2023 16:41 — 👍 14 🔁 5 💬 3 📌 2
Stem cell and developmental biologist. Modeling embryo development. Assistant Professor @Westlake University.
https://en.westlake.edu.cn/faculty/lizhong-liu.html
Postdoc fellow in the Plath lab at UCLA | Alum Cambridge University & Babraham Institute | Researching epigenetic changes during human embryonic development 🇬🇧🏳️🌈
Developmental biologist and Executive Editor of Development @dev-journal.bsky.social. Part of the Node @the-node.bsky.social and The Company of Biologists @biologists.bsky.social. Own views.
Stem Cells, Embryo Models, and Interspecies Chimeras. Weird is the new normal!
Stem cells and development, quantitative biology.
Assistant Professor of Biomolecular Engineering at UCSC shariatilab.sites.ucsc.edu
assisant prof @mcdb-yale.bsky.social / investigator @wutsaiyale.bsky.social
using tools from synthetic biology and physics to decode and control how cells build tissues and organisms
Penn MD/PhD student.
Runner when bones work
Reviews Editor at Development. Natural and Social Scientist in Stem Cell and Developmental Biology. Formerly reNEW (University of Copenhagen), @edinuni-irr.bsky.social and @oxforddpag.bsky.social. Views my own.
Reprogramming cells for fun and to create embryo models in the tropical Greater Bay Area
https://www.jose-silva-lab.com/ 🇵🇹🇨🇳🇬🇧
Guangzhou/Cantāo 🌴☀️, China中国
Biologist @hubrechtinstitute.bsky.social interested in signalling dynamics, development, tissue homeostasis and microfluidics | Formerly MPI Biochemistry, Biocenter Basel and Embl Heidelberg | Mother of 2 | Posts are my own.
Utrecht | sonnenlab.org
Senior Editor at @natcellbio.nature.com: #stemcells & development #disease #preclinical & #clinical studies | proud scientist | 🏳️🌈 non-binary, antisexist & liberal | 🍫 📚 🐶 🧑🍳 |📍Berlin | views are his own | call him Stelios
Genetics professor at Harvard Medical School. Interested in RNA life cycles and genome organization across the cell, from the nucleus to mitochondria.
Professor, Department of Medicine and McDonell Genome Institute @ Washington University. Specializing in Bioinformatics, Genomics, and Cancer. http://griffithlab.org/
Assistant Professor of Genetics and Core Member of the Epigenetics Institute at UPenn | T2T, HPRC, and HGSVC member | Loves genomics, epigenomics, and synthetic biology | logsdonlab.com
Assistant Professor, BME, Columbia University
Computational Biologist, Head of research @ebi.embl.org, part-time
Heidelberg University, codirector DREAM challenges. For group's activities, see @saezlab.bsky.social
Quantitative stem cell biologist and group leader @TheCrick Institute. Cell decision-making. Cell fate and cell division. @Stanford, @EMBL alumna. ENTP. Mum. Diver.
Scientist and mother…passionate about traveling and embracing challenges.
Dedicated to unraveling the mysteries of multicellular systems and stem cell biology. Combining experimental and theoretical approaches to understand the dynamics of cell behaviour
See https://tenuretracker.info/ for the most complete & worldwide overview of open postdoc, tenure track, (junior) professor, and lecturer positions.
Visit the site to search for specific positions and subscribe to email notifications