This #microscopymonday is a cleaning day. Watch how a #microglia cell mops up and phagozytoses debris and junk in a very messy neuronal co-culture. BTW, it was a proper cleaning job → the neurons made it and formed nice networks
Phase contrast imaging started at DIV1. One frame per 20 min.
12.10.2025 16:32 — 👍 24 🔁 8 💬 2 📌 0

Bioimage Analysis Specialist | King's College London
BioImage analysis friends - King's are recruiting for a full-time, permanent facility position! Come and work with fun microscopes and fun people (and me!) - please share! www.kcl.ac.uk/jobs/126345-...
29.09.2025 09:48 — 👍 61 🔁 80 💬 0 📌 0
Reconstructing long-range axons from dense brain images is tough.
This study introduces a novel method that separates axon identification from global statistical rules, showing big improvements over existing tools for mapping neuronal projections.
buff.ly/OBKr9cM
07.09.2025 10:01 — 👍 23 🔁 8 💬 0 📌 1

Figure legend: Categories and capabilities of smart microscopy systems integrating real-time image analysis and feedback control. Top: Smart microscopy workflows can be classified based on the driving logic behind decision-making: Event-driven (reacting to rare biological events), Outcome-driven (using feedback-control to steer biological systems toward a desired state), Quality-driven (optimizing signal quality or imaging metrics), and Information-driven (guided by models that predict which measurements/perturbations will yield the most informative data). Middle: Central feedback loop between the microscope and an image analysis system, which continuously
exchanges images and commands to guide acquisition dynamically. Bottom: Key control actions enabled by smart microscopy: adjusting imaging modality (e.g. switching from brightfield to fluorescence, adjusting sampling rate), repositioning the field of view (e.g. tracking, drift correction), optimizing acquisition settings (e.g. adaptive optics),
and performing photomanipulation (e.g. FRAP, ablation, optogenetics).
🔬🧠 Our paper on smart microscopy & the issue of interoperability! www.biorxiv.org/content/10.1... LONG THREAD WARNING: Smart microscopy uses real-time image analysis to automatically guide the acquisition or perturbation of the sample (closed feedback-control loop). Many applications exist:
21.08.2025 14:21 — 👍 36 🔁 23 💬 2 📌 1
Automated optogenetic control of hundreds of cells in parallel. Each cell is individually steered, collectively acting as a "tissue printer". Preprint & code out! www.biorxiv.org/content/10.1...
21.08.2025 20:16 — 👍 104 🔁 38 💬 6 📌 2

Our bryozoan paper is out in @genomeresearch.bsky.social! Led by @tomlewin.bsky.social, we report genome shuffling across the phylum and synteny evidence for the Lophophorata hypothesis. Grateful to John Bishop @thembauk.bsky.social and the DToL project for sharing openly!
doi.org/10.1101/gr.2...
07.01.2025 06:20 — 👍 41 🔁 13 💬 1 📌 0
FlyBase:Contribute to FlyBase - FlyBase Wiki
FlyBase needs your help! We ask that European labs continue to contribute to Cambridge, UK FlyBase, whereas US and other non-European labs can contribute to US FlyBase. For more information and how to donate: wiki.flybase.org/wiki/FlyBase...
15.08.2025 12:45 — 👍 128 🔁 158 💬 3 📌 26
A reminder you/your lab can support FlyBase at Cambridge through the following link. Every bit helps. Please share if you yourself can't donate.
www.philanthropy.cam.ac.uk/give-to-camb...
14.08.2025 05:25 — 👍 79 🔁 110 💬 1 📌 5
Reminder that all three books I've co-authored are freely available online for non-commercial use (and the fourth will be, too)
11.08.2025 17:44 — 👍 154 🔁 49 💬 4 📌 1
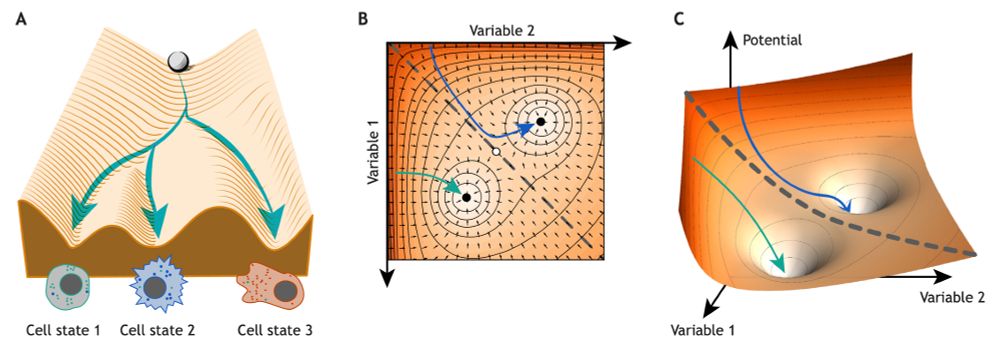
Adapted from Fig. 1 from Kadiyala et al 2025.
Introducing five concepts from dynamical systems to decode developmental regulatory mechanisms, have a read! @perez-carrasco.bsky.social @roederlab.bsky.social @mpipz.bsky.social This effort started in a morphogenesis meeting @kitp-ucsb.bsky.social in 2023. journals.biologists.com/dev/article/...
04.08.2025 06:42 — 👍 69 🔁 35 💬 1 📌 2
I read and re-read the paper and talked to some on the experienced editorial staff.
My conclusion was that there were not sufficient reasons to retract the paper. The evidence was much weaker than was needed to fully support the claims. but this was true to some degree for many papers.
9/n
27.07.2025 11:48 — 👍 37 🔁 2 💬 6 📌 3
At this point, I might as well --
Here's an infographic showing different ways to include age as a predictor. The top shows two extremes, just as a plain old numerical predictor (imposes linear trajectory) vs. categorical predictor (imposes nothing whatsoever). And then three solutions in between!
16.07.2025 12:33 — 👍 211 🔁 47 💬 22 📌 1


The "reproducibility crisis" in science constantly makes headlines. Repro efforts are often limited. What if you could assess reproducibility of an entire field?
That's what @brunolemaitre.bsky.social et al. have done. Fly immunity is highly replicable & offers lessons for #metascience
A 🧵 1/n
10.07.2025 08:21 — 👍 318 🔁 173 💬 10 📌 18

Some rough design guidelines*
1. Match effectiveness with importance
2. Avoid ambiguity
3. Locality is king / eyes beat memory
4. Establish viewing order
5. Layer, layer, layer
6. When in doubt, grid
7. Treat visual attributes like adjectives
* These guidelines are drawn largely from my experience + personal preferences + the literature.
Design is messy, these are not perfect, others will disagree with me, etc. Caveat emptor.
These are the design guidelines I teach. From halfway through this deck: www.mjskay.com/presentation...
These days (for academic vis) I'd add "ensure natural visual operations correspond to meaningful operations in data space". Need to make slides for that.
03.07.2025 18:21 — 👍 37 🔁 10 💬 4 📌 3
A checklist for designing and improving the visualization of scientific data
Nature Cell Biology - Creating clear and engaging scientific figures is crucial to communicate complex data. In this Comment, I condense principles from design, visual perception and data...
I gave in! After students asking for it, I now made a simple figure design checklist.
To help all scientists w/o graphic skills create clear, accessible, and truthful charts!
-> Out in @nature Cell Biology: rdcu.be/erwl4
#DataVisualization #PhD #SciComm
Thx for review @bethcimini.bsky.social + 2
18.06.2025 08:33 — 👍 294 🔁 116 💬 15 📌 3
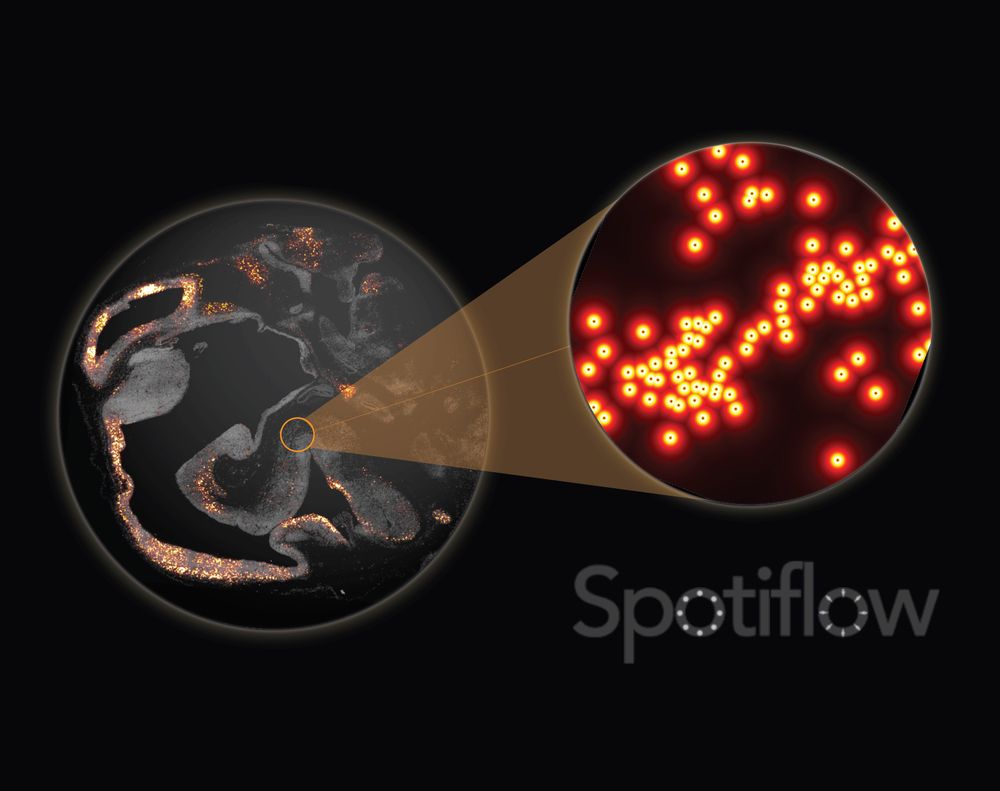
Out today in @natmethods.nature.com : Spotiflow, our transcript localization method for imaging-based spatial transcriptomics. Led by amazing PhD student @albertdm.bsky.social, joint work w @gioelelamanno.bsky.social at EPFL / @scadsai.bsky.social
www.nature.com/articles/s41...
rdcu.be/epIB7
06.06.2025 19:05 — 👍 109 🔁 37 💬 9 📌 2
Drosophila Genetic Database
The Drosophila Genetic Database, FlyBase, is on the brink of collapse due to the sudden termination of the FlyBase NIH grant, which includes salaries for 5 literature curators based at the University ...
#drosophila @flybase.bsky.social request emergency funding:
"As it stands, by the end of July, 2025, there will be no future updates to FlyBase, and in the worst case scenario access to the website will also be lost" => please donate!
www.philanthropy.cam.ac.uk/give-to-camb...
03.06.2025 16:15 — 👍 81 🔁 102 💬 3 📌 17
ok stats experts!
if we have measures in a timecourse, how would you capture whether different groups show a different pattern?
i guess the simple/obvious thing is t test on a given timepoint but this throws away all the info from multiple timepoints...
any link/tutorial you like? (pls rski)
02.06.2025 16:49 — 👍 14 🔁 9 💬 12 📌 0

Toward a probabilistic definition of neural cell types
A classical view of cell type relies on a definite set of stable properties that are critical for brain functions. Single-cell technologies led to an …
@maheandria.bsky.social and I challenge the notion of a static and deterministic definition of a (neural) cell type and argue for its replacement by a multi parametric probabilistic definition in this perspective
sciencedirect.com/science/arti...
06.05.2025 12:49 — 👍 10 🔁 3 💬 0 📌 2
Introducing warpfield, an open source Python library for GPU-accelerated non-rigid 3D registration. Warps and aligns gigavoxel volumes within seconds (not hours). For 3D microscopy, region-to-region and cell-to-cell matching.
A collaboration with @mh123.bsky.social 🚀
github.com/danionella/w...
12.05.2025 05:25 — 👍 195 🔁 54 💬 7 📌 3
Get Better: R for cell biologists – quantixed
An idea for a workshop that you can run (or take yourself!) to teach "R for cell biologists". 🧪
quantixed.org/2025/01/20/g...
20.01.2025 14:49 — 👍 39 🔁 8 💬 6 📌 1
"Die Neurobiologie der Künstlichen Intelligenz": Vortrag im Rahmen der Dahlemer Wissenschaftsgespräche am 15. Januar um 18 Uhr. Eintritt frei.
📍Freie Universität Berlin, Forschungsbau SupraFAB, Altensteinstraße 23a, 14195 Berlin, Meeting Point Raum 201
▶️Infos: www.fu-berlin.de/presse/infor...
09.01.2025 12:22 — 👍 3 🔁 2 💬 0 📌 0
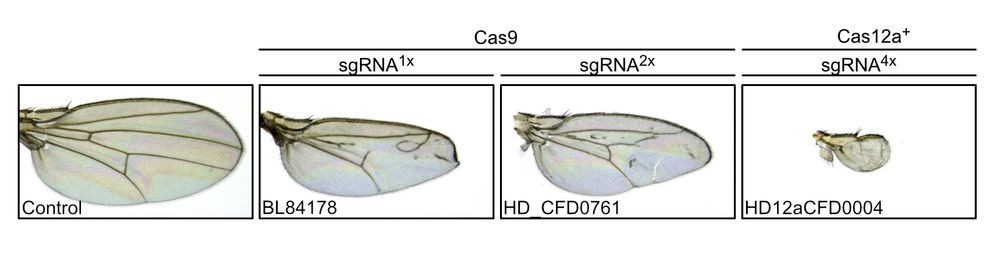
Images of Drosophila wings. Control wings expressing CRISPR components exclusively in the germline have normal morphology. Targeting the central hedgehog signaling component smoothened with Cas9 and either one or two sgRNAs results in wings of reduced size and missing and ectopic vein tissue (middle panels). Targeting smoothened with Cas12a+ and four sgRNAs results in very small wings without veins.
The power of Cas12a-enabled sgRNA multiplexing for gene disruption.
Today’s example: smoothened
1/3
#CRISPR #Drosophila
07.01.2025 21:25 — 👍 16 🔁 1 💬 2 📌 0
There are now 24 step-by-step guides, explaining how to create these ⬇️ data visualizations with R/ggplot2 - I hope this is a useful resource. Find them all here: joachimgoedhart.github.io/DataViz-prot...
16.12.2024 09:25 — 👍 547 🔁 192 💬 17 📌 14
Captain of the Good Team. Artist, Visionary, None of your Businessman, and paperback writer for kids.
Neuroscientist and glial aficionado at NYU Grossman School of Medicine/NYU Langone Health in NYC. Posts in my individual/personal capacity.
My lab is full of awesome people doing amazing stuff - check them out: www.liddelowlab.com
he/him
cell biologist heavily exposed to mechanics, development, and synthetic biology | postdoc @OatesLab prev. PhD @HeisenbergCPLab BS @BilkentUniv
A better place for my shit cartoons about the TERF WARS
See https://tenuretracker.info/ for the most complete & worldwide overview of open postdoc, tenure track, (junior) professor, and lecturer positions.
Visit the site to search for specific positions and subscribe to email notifications
Menswear writer. Editor at Put This On. Words at The New York Times, The Washington Post, The Financial Times, Esquire, and Mr. Porter.
If you have a style question, search:
https://dieworkwear.com/ | https://putthison.com/start-here/
🇨🇭 in 🇦🇹 Life beyond dogma! Free-floating systems thinker & natural philosopher. Antifragilist extemporanian metamodernist. Open science, society & living […]
[bridged from https://spore.social/@yoginho on the fediverse by https://fed.brid.gy/ ]
Just an ole t-test in a GLMM world. #statistics #healthcare #rstats
Professor of Biophysics -
@ISTAustria Using physics to understand biological questions, such as how embryos grow!
Assistant Professor @biozentrum.unibas.ch • Theoretical biophysics • Postdoc ISTAustria, PhD LMU Munich, MSc Cambridge University
www.biozentrum.unibas.ch/brueckner
Scientist interested in the brain, synapses, organelles, lipids, and much more
RE4GREEN is a three-year Horizon Europe project creating a framework to support the green transition by addressing ethics and integrity in research with environmental impacts and in climate-focused knowledge and technologies.
Decoding how the gut thinks 🦠🪱🧠💪
Neuroscientist with interests in
#EnergyMetabolism #EntericNeurons #Fats
@crick.ac.uk @institutducerveau.bsky.social
Prof at University of Bern
#MAPK #Rho #optogenetics #biosensors #computer_vision
skiing, rock/ice/mountain climbing when I have time
Evolutionary biologist at Academia Sinica, Taiwan. We study the evolutionary genomics of marine invertebrates and use sequencing approaches to explore their biodiversity. More at: https://sgel.biodiv.tw/
Biophysicist, Group Leader at MPIPZ. Studying the multicellular dynamics in plants, combining theory and experiments. Also, fascinated by music and dance.
Research laboratory of PI Adrienne Roeder in the Weill Institute of Cell and Molecular Biology and the School of Integrative Plant Science at Cornell Univ. We study morphogenesis and pattern formation in Arabidopsis and diatoms.
Systems Biology researcher at Imperial College interested in nonlinear & stochastic mechanisms. Tolerable clarinet player & painter http://instagram.com/twopiruben 🏳️🌈
Open-source statistics program that is free, friendly, & flexible.
Armed with an easy-to-use GUI, we enable both classical & Bayesian analyses.
https://jasp-stats.org/
Testimonials: https://jasp-stats.org/alltestimonials/