Scientific Advisory Board Meets at the Helmholtz Institute Würzburg
Two days of intensive scientific exchange
Two days of inspiring exchange 🧬🤝✨
On February 10 and 11, our Scientific Advisory Board came together to review the scientific orientation and performance of #HIRI. We are grateful for the insightful discussions and valuable feedback.
👉 Read more: www.helmholtz-hiri.de/en/newsroom/...
12.02.2026 15:15 — 👍 7 🔁 1 💬 0 📌 0

Thrilled to announce that I’ve just opened my research lab at @helmholtz-hiri.bsky.social !
A huge thank you to my mentor Roi Avraham (and lab members!) for the incredible training and support that made this possible.
06.02.2026 12:34 — 👍 17 🔁 12 💬 2 📌 1
Do you want to understand how RNA #splicing decisions are made? We‘ve got a PhD position open for you (see 👇👇)!!
Please share #RNAsky and RT!
tinyurl.com/4ztu9cb9
11.02.2026 08:20 — 👍 9 🔁 12 💬 0 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
📢 We are hiring 📢 1st round of recruiting for our ERC project BacImmune-Decode! @erc.europa.eu
We are looking for a postdoc for wet-lab work on regulation of phage defence systems in bacteria using high-throughput microbial genetics. Check it out: lnkd.in/es7AE968
#Hiring #Postdoc #Microbiology
14.01.2026 08:55 — 👍 13 🔁 19 💬 0 📌 0

Uropathogenic Escherichia coli invade luminal prostate cells via FimH–PPAP receptor binding - Nature Microbiology
Uropathogenic Escherichia coli infection of a murine prostate organoid model reveals a bacterial FimH–host prostatic acid phosphatase adhesin-receptor interaction enabling invasion and replication wit...
Our first paper is out today in @natmicrobiol.nature.com!
We introduce a prostate organoid model and show that UPEC invades prostate cells via FimH binding to the prostate-specific protein PAPP. A step toward targeted therapies against bacterial prostatitis. 🎉
#UTI #UPEC #Organoids #AMR
08.01.2026 11:12 — 👍 34 🔁 19 💬 2 📌 2

Intramacrophage RIL-seq uncovers an RNA antagonist of the Salmonella virulence-associated small RNA PinT
Abstract. Salmonella virulence chiefly relies upon two major pathogenicity islands, SPI-1 and SPI-2, which enable host cell invasion and intracellular surv
RIL-seq boosts the study of sRNAs. Here comes its first application to intracellular bacteria. Plus, 10 years after its characterization, we now present an RNA sponge of Salmonella PinT.
Congrats to @kooshapour.bsky.social & great collaboration w/ @jorg-vogel-lab.bsky.social!
doi.org/10.1093/nar/...
27.12.2025 14:17 — 👍 30 🔁 17 💬 1 📌 0

🎓 Congrats to Elise Bornet, who completed her PhD this week! Her courageous work dissected morphological heterogeneity in predominant human gut commensals, and yielded an ultra-sensitive transcriptomics pipeline to sequence the RNA content of single Bacteroides cells. doi.org/10.1016/j.ce...
20.12.2025 10:06 — 👍 6 🔁 0 💬 0 📌 0

Grateful to be among the 25 inspiring women featured in @zeit.de as part of the #Zia fellowship for Visible Women in Science. Thanks to @uni-wuerzburg.de for their support and to everyone involved for making scientific work by women more visible. Honored and proud to be part of this initiative ☺️.
19.12.2025 15:24 — 👍 7 🔁 3 💬 0 📌 0

How does a bacterial small RNA evolve new functions?
Our new study reveals the stepwise evolutionary journey of UhpU, a 3′UTR-derived sRNA in E. coli and relatives. It evolved from a metabolically-focused regulator to one with expanded targets and biogenesis manner.
www.pnas.org/doi/10.1073/...
04.12.2025 19:50 — 👍 15 🔁 12 💬 2 📌 0
Job offer for a PhD student position at the Chair of Microbiology
🔊 Interested in doing a PhD on RNA-binding proteins in an abundant microbiota species and becoming a member of the German Priority Programme “Illuminating Gene Functions in the Human Gut Microbiome”? Apply here: www.uni-wuerzburg.de/karriere/sin...
01.12.2025 09:20 — 👍 7 🔁 15 💬 0 📌 1

De-DUFing the DUFs 🧩 @franznarberhaus.bsky.social lab uncovers how small DUF1127 proteins regulate #phosphate uptake by binding the sensor kinase PhoR. Their conserved role from Agrobacterium to E. coli highlights how even small DUFs can shape bacterial physiology 🦠
buff.ly/jJd9Eho
21.10.2025 07:01 — 👍 21 🔁 14 💬 1 📌 0

Excited to share our preprint led by Carlos Voogdt et al
We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...
13.10.2025 07:47 — 👍 85 🔁 45 💬 2 📌 4

Congrats to Ann-Sophie Rüttiger who successfully defended her PhD this week! 🎓
She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).
25.09.2025 10:12 — 👍 13 🔁 3 💬 0 📌 0

Looking for a new approach to studying or eliminating phages? Check out our study introducing anti-phage ASOs (antisense oligos) out in @Nature today. nature.com/articles/s4158…
10.09.2025 15:40 — 👍 133 🔁 65 💬 4 📌 2

Starting the meeting microbes and RNA- Gerhart on stage - lifetime achievement Lecture - congrats Gerhart!
01.09.2025 14:19 — 👍 5 🔁 1 💬 2 📌 0
A Salmonella T3SS-2 mutant grows fine in spleen macrophages, contradicting tissue culture dogma (PMID: 23236281). This observation was largely ignored, but adding certain carbon sources rescues growth in cultured macrophages, hinting that T3SS-2 may be doing something entirely different in vivo.
09.08.2025 00:18 — 👍 25 🔁 11 💬 0 📌 1
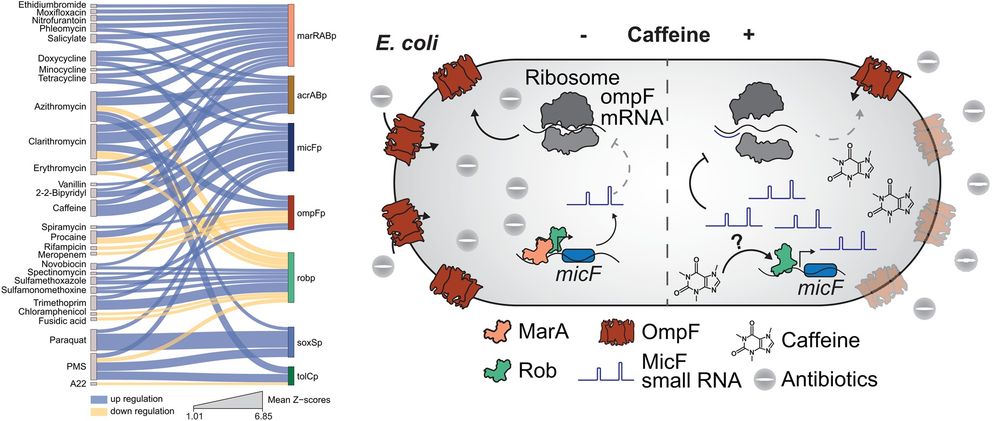
Left: Compound-promoter interaction network. Fifty-three significant compound-promoter interactions are shown as edges in a Sankey diagram connecting the compounds (left, source nodes) to the promoters (right, target nodes). Edge thickness represents mean Z-scores (n = 8), while node size represents the total number of interactions. Right: Proposed model for the molecular mechanism of caffeine-ciprofloxacin antagonism. Caffeine triggers expression of MicF small RNA in a Rob-dependent manner, which then binds to the 5′-UTR of ompF mRNA to inhibit and decrease OmpF protein levels. This ultimately prevents ciprofloxacin from entering the cell, resulting in caffeine-ciprofloxacin antagonism
How are bacterial transport machineries regulated by external cues? @brochadolab.bsky.social &co identify environmental chemicals that regulate #transporter transcription, identifying the TF Rob as a key modulator of #EffluxPump expression in #Ecoli #AMR @plosbiology.org 🧪 plos.io/46pUM9Q
23.07.2025 09:20 — 👍 36 🔁 13 💬 1 📌 0
New preprint! We characterise the small, regulatory RNA Arp which controls DNA uptake and twitching motilty in A. baumannii. Led by our Dr Fergal Hamrock @hamrockfergal.bsky.social and in collaboration with @westermannlab.bsky.social and Mike Gebhardt's lab!
21.07.2025 14:56 — 👍 3 🔁 6 💬 0 📌 1
Congrats to Gianluca Prezza (@gprezza.bsky.social) for reveiving the 2025 PhD Award from the Friends of the Helmholtz Centre for Infection Research (HZI)!👏 The award honours excellent PhD theses from the field of life sciences at HZI partner universities. His thesis focused Bacteroides' RNA biology.
11.07.2025 09:35 — 👍 10 🔁 3 💬 0 📌 1
Thank you to all students and to the GSLS for this award! It means a lot to me! (-AW)
07.07.2025 07:46 — 👍 10 🔁 0 💬 1 📌 0
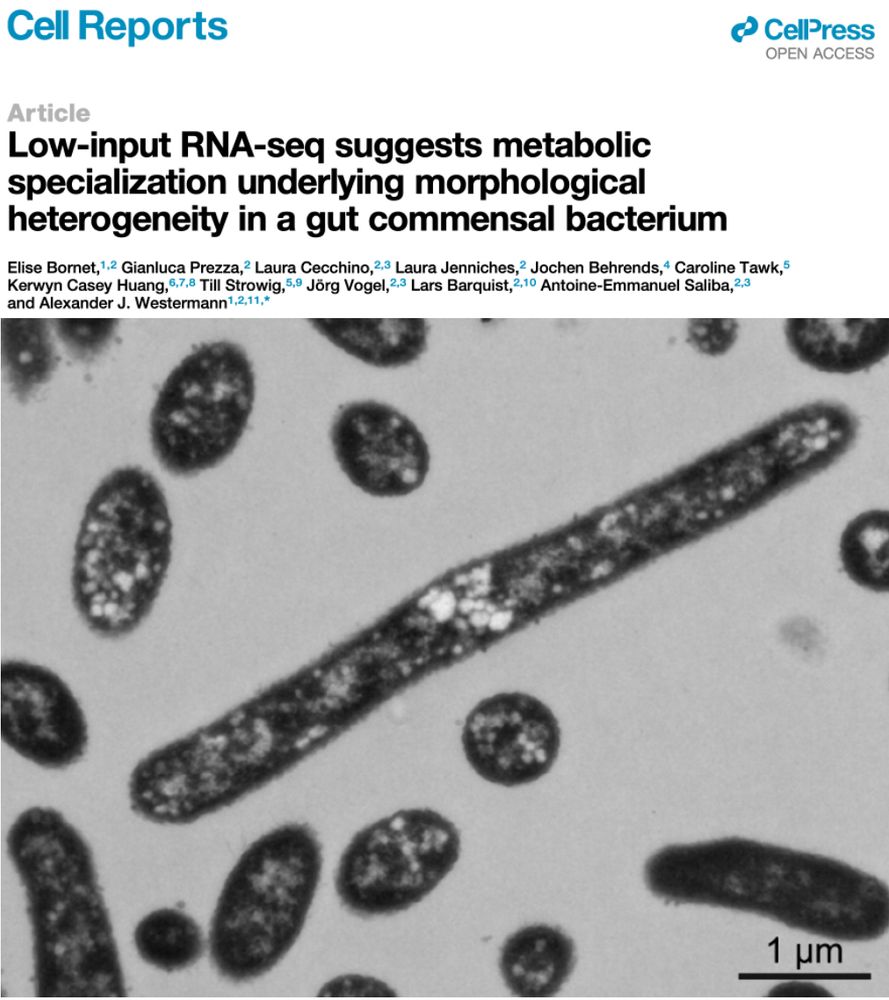
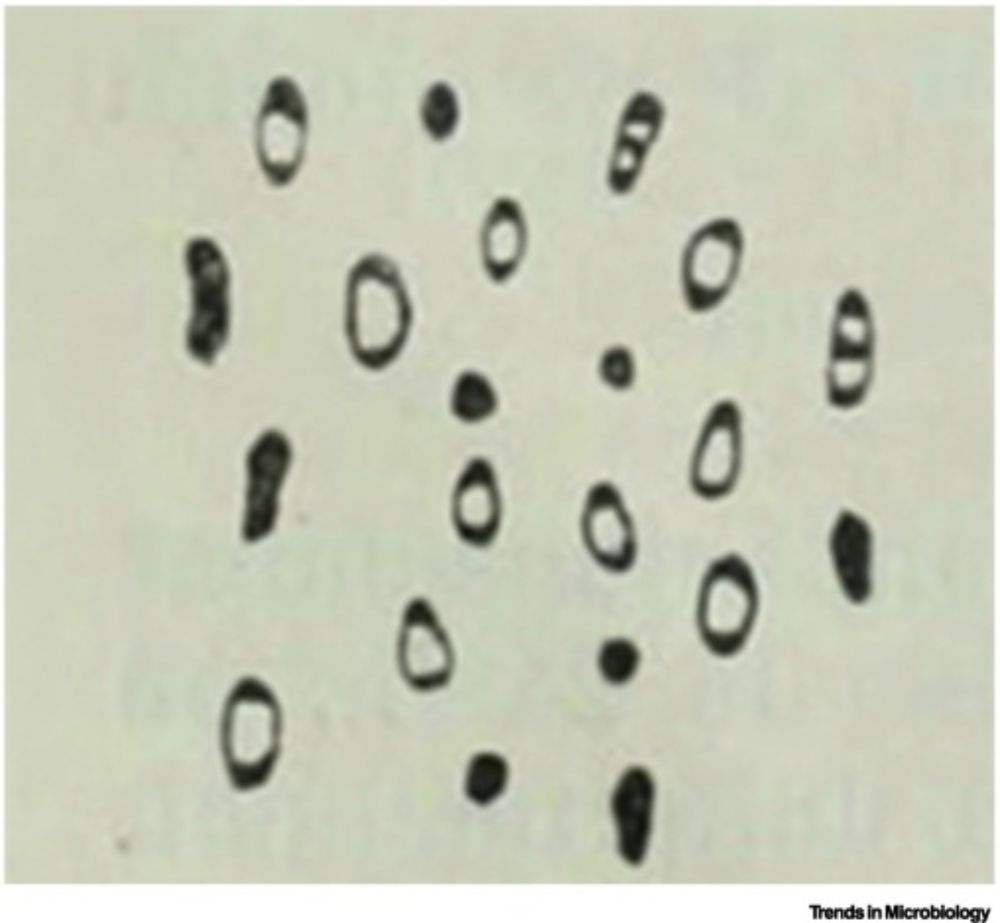
𝔟α𝐜𝐓𝕖Ř𝒾ⒶĹ SHⓐ𝓹Ẹ 丂ħ𝓲ƒ𝓉ⒺⓇร 🦠
Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...
18.06.2025 01:02 — 👍 14 🔁 8 💬 0 📌 0
Dad, Teacher, Artist, Gardener and Scientist.
Change is the only permanent thing in the world. This moment will pass. It has to.
Ps.
All I post is my personal opinion
BIOspektrum - Das Magazin für Biowissenschaften * Neue Themen aus der Wissenschaft * Nachrichten aus der Scientific Community
http://www.biospektrum.de
Biologist and PhD in Chemistry from 🇦🇷 | Passionate about #microbes | #MSCActions and #EMBL alumni of @zimmermannlab.bsky.social | Proud #mom of 2
Microbiome research, Microbial communities, evolution, infection and diet. Postdoc at Helmholtz HZI. Researcher at Yale MSI Goodman lab. PhD Würzburg Vogel lab. AUB Alumnus.
We use high-throughput MS, bacterial genetics, gnotobiology & computational models to study metabolic interactions in microbial ecosystems
@EMBL
Homepage: https://www.embl.org/groups/zimmermann/
Uncovering the untapped genetic potential of the human gut microbiome: a DFG-funded Priority Programme decoding unknown gene functions and their pathway organisation. Learn more at www.spp2474.de
assistant professor at georgetown | polymicrobial interactions | bacterial physiology | science educator | views my own
www.zarrellalab.org
Lecturer/Assist. Prof. in Bacteriology @ University of Glasgow | de novo evolution of bacterial gene regulatory networks |
✌️❤️ RNA | she/her
#Science #PhD #PhDlife #StructuredTraining #UNIWürzburg
@uniwuerzburg.bsky.social
@drcwuerzburg.bsky.social
German scientific society for all those interested in biochemistry, molecular biology, biomedicine, biotechnology, cell biology, genetics...
RNA biology | Gut microbes
https://ibb.edu.pl/en/laboratory/daniel-ryan-phd/
Leader of the Nanoscale Bacteriology research group at the Rudolf Virchow Center @ JMU Würzburg. Revealing the inner life of #bacteria using #SuperResolutionMicroscopy.
Human immunology through the RNA lens. Focused on macrophages, lncRNAs & systems immunology. Professor for RNA- and infection-biology @unimarburg.bsky.social. Web: http://rna-lab.org/
Microbiologist with a particular interest for small RNAs and bacterial persistence ! Alumni of the Vogel and Helaine labs. Researcher in Rennes (Inserm U1230).
Nitrogenase, Biotic and Artificial Metalloenzymes, CO2 and N2 Fixation, Emmy Noether Research Group Leader at the Max Planck Institute for Terrestrial Microbiology: https://www.mpi-marburg.mpg.de/859249/Johannes-Rebelein
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Scientific writer @helmholtz-hiri.bsky.social and editor at Life Science Editors (https://www.lifescienceeditors.com/)
RNA, microbiology, infectious disease; Helmholtz Institute for RNA-based Infection Research & University of Würzburg, Germany
www.helmholtz-hiri.de