I talk a lot about Rust for building high-perf (& even non-perf critical) software, & scientific software in particular. I often discuss what's interesting to me, but wanted to offer the chance to those interested for me to answer their questions about Rust in science. Fire away with questions!🧬🖥️
03.08.2025 18:12 — 👍 19 🔁 11 💬 4 📌 1
Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
Know an Evolutionary Genomicist looking for a faculty position? Join us in EEB @utknoxville.bsky.social Position is open for studying any organism, but personally I have some botany bias. 🌱 Apply before Sept 19 for full consideration. apply.interfolio.com/170735 Please share widely. Thanks!
02.08.2025 16:16 — 👍 53 🔁 74 💬 2 📌 0

I confirm your opinion
02.08.2025 22:12 — 👍 1 🔁 0 💬 0 📌 0
My first first author manuscript has posted! I’m very excited to share SWIF-TE with yall. It is a fast, memory efficient tool to identify novel TE insertions from short-read data. 🧬 #TEworldwide #transposons
02.08.2025 18:42 — 👍 64 🔁 17 💬 1 📌 2
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
02.08.2025 20:39 — 👍 13 🔁 7 💬 2 📌 0
Watching @jonhoo.eu's vibe-coding stream exactly matches my personal experience using agentic AI for programming. That is, it's fine for things that are basiclly pattern matching, but fails quite badly (i.e. is slower than just doing it manually) for changes that require reasonable thought.
02.08.2025 17:09 — 👍 4 🔁 1 💬 0 📌 0
![\toprule
& & \multicolumn{4}{c}{CPU time / h} & & \multicolumn{4}{c}{Max RSS\tnote{a}{ } / GB} \\
\cmidrule(lr){3-6} \cmidrule(lr){8-11}
Read length / bp & & 24 & 100 & 200 & 24--200 & & 24 & 100 & 200 & 24--200\\
\midrule
Newmap & & 4 & 9 & 16 & 150.5 & & 7.6 & 7.6 & 7.6 & 7.6 \\
Genmap & & 2 & 1 & 1 & 275 & & 13.1 & 13.1 & 13.1 & 13.1 \\
Umap & & 31 & 42 & 57 & >4752\tnote{b} & & 7.8 & 7.5 & 5.9 & ${\sim}7.8$\tnote{c}\\
\bottomrule
\end{tabular}
\begin{tablenotes}
\item[a] Maximum resident set size.
\item[b] Estimate from a linear extrapolation of \qty{24}{\basepair} CPU time.
\item[c] Estimate from the largest maximum RSS found across all Umap benchmarks.
\end{tablenotes}](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:y6iikj3lebwl7k5cyi2fbahv/bafkreieliq6x7p4h6dqvxsgvzlitbptse3gl55qayic6qn3wqacsakxsd4@jpeg)
\toprule
& & \multicolumn{4}{c}{CPU time / h} & & \multicolumn{4}{c}{Max RSS\tnote{a}{ } / GB} \\
\cmidrule(lr){3-6} \cmidrule(lr){8-11}
Read length / bp & & 24 & 100 & 200 & 24--200 & & 24 & 100 & 200 & 24--200\\
\midrule
Newmap & & 4 & 9 & 16 & 150.5 & & 7.6 & 7.6 & 7.6 & 7.6 \\
Genmap & & 2 & 1 & 1 & 275 & & 13.1 & 13.1 & 13.1 & 13.1 \\
Umap & & 31 & 42 & 57 & >4752\tnote{b} & & 7.8 & 7.5 & 5.9 & ${\sim}7.8$\tnote{c}\\
\bottomrule
\end{tabular}
\begin{tablenotes}
\item[a] Maximum resident set size.
\item[b] Estimate from a linear extrapolation of \qty{24}{\basepair} CPU time.
\item[c] Estimate from the largest maximum RSS found across all Umap benchmarks.
\end{tablenotes}
The new version is built on @wheelerlab.org's AvxWindowedFMindex and some other tricks we came up with, and is *so fast*.
01.08.2025 19:00 — 👍 4 🔁 1 💬 0 📌 0
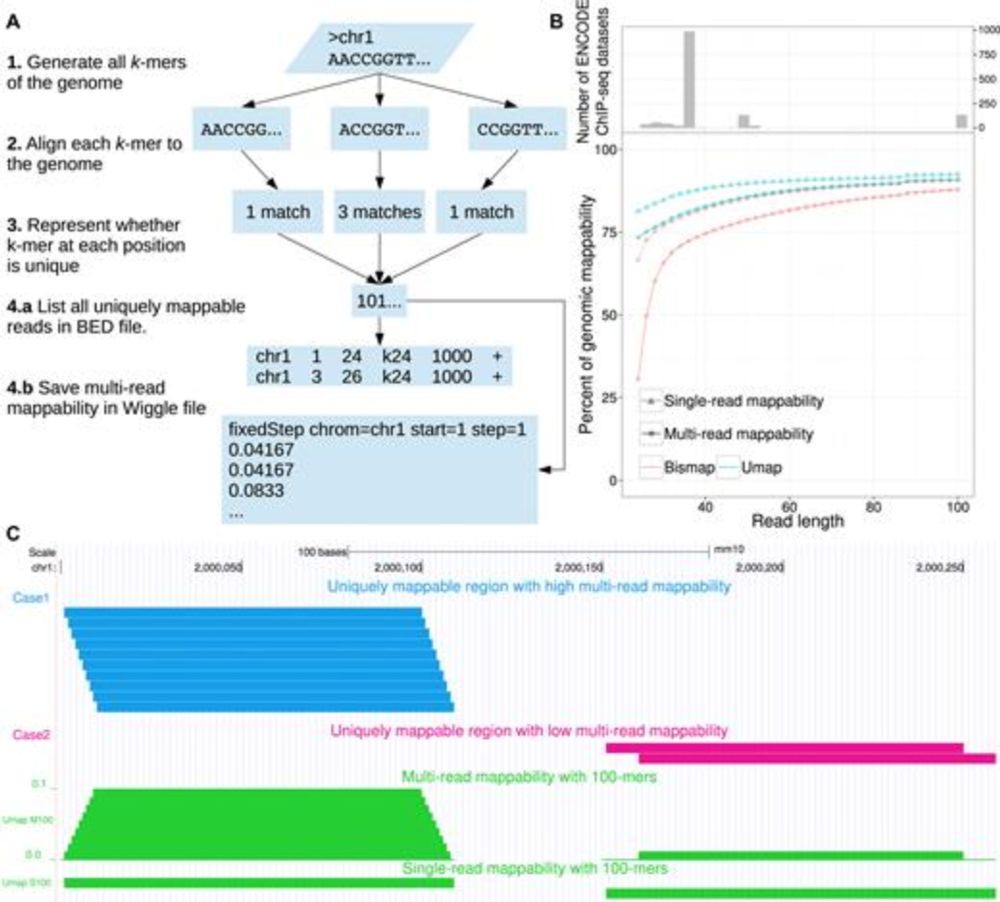
Umap and Bismap: quantifying genome and methylome mappability
Abstract. Short-read sequencing enables assessment of genetic and biochemical traits of individual genomic regions, such as the location of genetic variati
We are creating new software to quantify genome mappability, a successor to Umap, our previous work described in the linked paper. What standard read lengths should we provide in BED/Wiggle format? Previous was 24/36/50/100 bp. academic.oup.com/nar/article/...
01.08.2025 18:51 — 👍 26 🔁 4 💬 5 📌 0

Really important points by @alexbateman1.bsky.social on the importance of curation, addressing some of the myths out there.
Deeply resonates with the commentary Paul Thomas and I wrote last year www.nature.com/articles/s41...
24.07.2025 08:13 — 👍 13 🔁 6 💬 0 📌 1
it's longer than the drive from ANY house in Missoula to the Missoula airport.
22.06.2025 17:20 — 👍 1 🔁 0 💬 1 📌 0
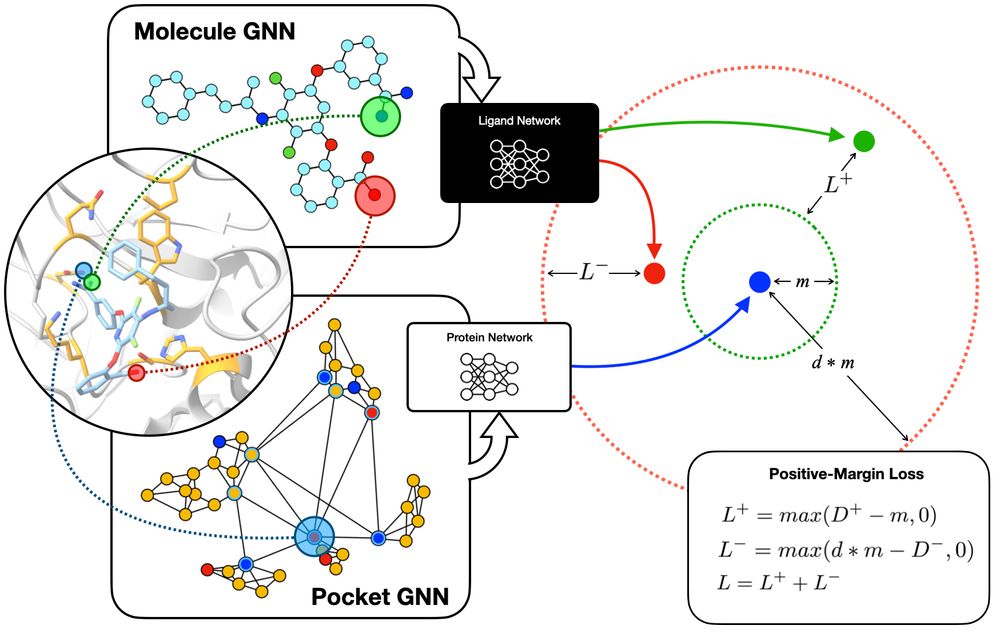
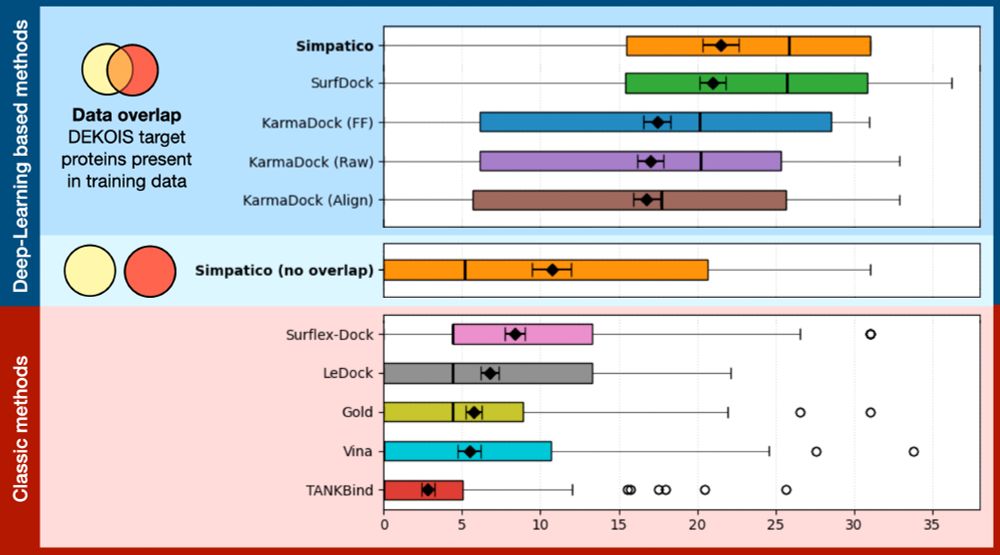
A Representation Learning approach to virtual ligand-screening. Produces good enrichment for active ligands, and can scan billions of candidate small molecules in a few hours..
Simpatico: www.biorxiv.org/content/10.1...
20.06.2025 14:47 — 👍 3 🔁 1 💬 0 📌 0
That is 54, 5-year NIH biomedical research grants.
Or 270 years worth of biomedical research.
10.06.2025 17:19 — 👍 109 🔁 54 💬 3 📌 0
YouTube video by wjteegarden
Now listen to this carefully, Norman: I am lying...
www.youtube.com/watch?v=4WRt...
10.06.2025 19:50 — 👍 1 🔁 0 💬 1 📌 0

Generated image of a very complicated multi-head candle, with all ends burning in a mighty conflagration. In the background (blurred), is a bewildered-looking man watching the candles burn.
I am definitely NOT burning the candle at both ends
09.06.2025 21:07 — 👍 1 🔁 0 💬 0 📌 0

Hello bluesky world! Newbee here! I have a postdoc position immediately available in my lab. It will focus on identifying high-quality transposons in many genomes and finding their impacts in evolution and traits. Most works, including EDTA2 development and annotation of 400+ genomes, are done! 1/n
10.04.2025 18:08 — 👍 25 🔁 22 💬 3 📌 2
@googleworkspace.bsky.social :
Is this ***yet another*** in an endless line of features that non-paying customers get to use, but I can't access because I have a paid workspace account? I'm running out of room to keep track of them all:
blog.google/products/map...
09.06.2025 04:09 — 👍 0 🔁 0 💬 0 📌 0
👀 👀 👀
07.06.2025 20:28 — 👍 0 🔁 0 💬 0 📌 0
Some of us look for ways to develop a joke over the course of a paragraph. Others just look for the quick win with a responsive quip.
But really: everybody's looking for something.
06.06.2025 22:44 — 👍 2 🔁 0 💬 1 📌 0
Very excited to share I’ll be starting my own group at University of Utah in the Department of Human Genetics in the new year! Reach out if you are interested! vollgerlab.com
05.06.2025 14:15 — 👍 62 🔁 11 💬 8 📌 4

Received on 05 February 2025;
Interesting read -- I'd have agreed whole-heartedly when it was written. Starting to feel a little dated:
04.06.2025 06:43 — 👍 2 🔁 0 💬 1 📌 0
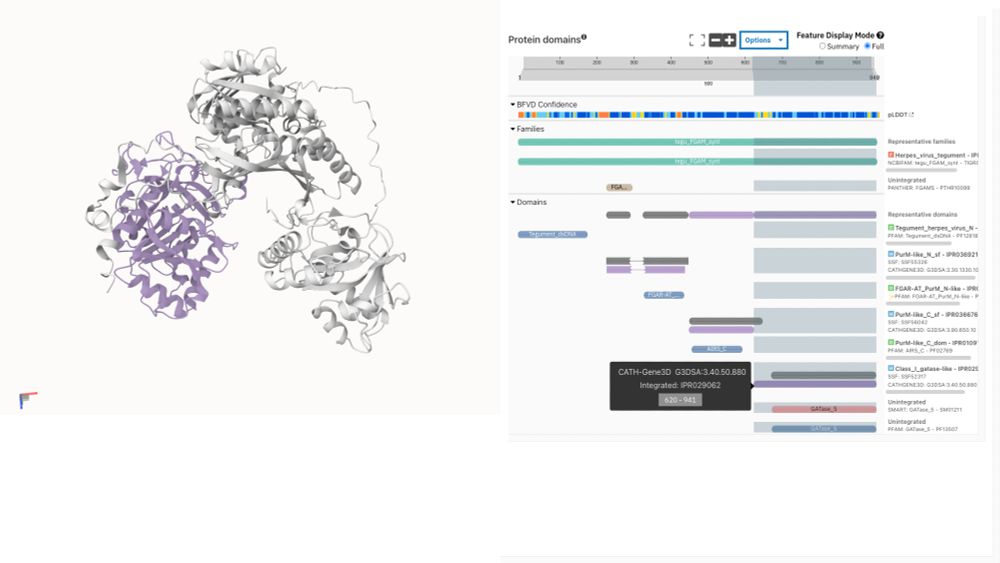
CATH-Gene3D annotation visualised on BFVD structure prediction of UniProt A0A075FDU7
🦠🧬Exciting news! BFVD predicted viral protein structures are now available in @InterProDB!
For the first time, explore InterPro functional annotations directly on 3D structures of viral proteins not covered by AlphaFold DB. This bridges a critical gap in structural virology research.
03.06.2025 12:47 — 👍 29 🔁 9 💬 2 📌 1
Ochman Lab: HOME
Howard's: evolution of microbial genes, genomes and communities. Can be experimental or computational. If interested, contact Howard Ochman, howard.ochman@austin.utexas.edu
Homepage: web.biosci.utexas.edu/ochman/index...
02.06.2025 20:26 — 👍 12 🔁 14 💬 0 📌 0
Nancy Moran Contact
An update on open postdoc positions, in my group and the adjacent group of Howard Ochman, here at UT-Austin. #SymbioSky
Mine: insect-bacterial symbiosis with emphasis on intracellular endosymbionts and molecular mechanisms.
web.biosci.utexas.edu/moran/contac...
02.06.2025 20:26 — 👍 16 🔁 23 💬 1 📌 2
mdrepo.org is still live and kicking, and still accepting new contributions.
02.06.2025 15:24 — 👍 1 🔁 0 💬 0 📌 1
Phage people: Rich Losick and I are combing the world looking for T4 rIIB mutant FC0 (also known as P13). FC0 was the starting point for Francis Crick's beautiful 1961 paper on the triplet nature of the genetic code. We want to sequence it. Anyone have it in an ancient stock collection?
01.06.2025 14:46 — 👍 64 🔁 79 💬 4 📌 5

A quote from Matthew 25:41–46: (The people on the left are the goats)
41 “Then he will say to those on his left, ‘Depart from me, you who are cursed, into the eternal fire prepared for the devil and his angels. 42 For I was hungry and you gave me nothing to eat, I was thirsty and you gave me nothing to drink, 43 I was a stranger and you did not invite me in, I needed clothes and you did not clothe me, I was sick and in prison and you did not look after me.’
44 “They also will answer, ‘Lord, when did we see you hungry or thirsty or a stranger or needing clothes or sick or in prison, and did not help you?’
45 “He will reply, ‘Truly I tell you, whatever you did not do for one of the least of these, you did not do for me.’
46 “Then they will go away to eternal punishment, but the righteous to eternal life.”
I guess she just really wants to be a GOAT (Matthew 25:31-46):
01.06.2025 00:08 — 👍 0 🔁 0 💬 0 📌 0
I was 100% certain that this was a deepfake, 'cause ... I mean, do I need to explain? But, nope, it's the real deal:
thehill.com/homenews/sen...
01.06.2025 00:00 — 👍 1 🔁 0 💬 1 📌 0
The European Nucleotide Archive offers open data archiving across species, platforms & applications. A globally comprehensive record & platform for data coordination
Asst. prof. @VUBrussel.bsky.social | Senior Postdoc in computational protein design and structural bioinformatics @VibLifeSciences.bsky.social VIB-VUB Center for Structural Biology, Brussels (Lab of @AVorobieva.bsky.social)
🌍Direct DNA/RNA analysis for anyone, anywhere
🧬Short to ultra-long reads in real-time for rapid insights
🔬Products for research use only
computational biologist. algorithms for genomics. worried about sustainability: from personal 2 country 2 planet. 🧪🧬| http://rivaslab.org
Spartan, Assistant Professor @OhioState. Trying to understand how plants co-evolve with TEs. Author of LTR_retriever, LAI, EDTA, panEDTA, TESS...
InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites.
ebi.ac.uk/interpro/
Research biologist and lover of nature, mostly bacteria, insects and (lately) birds
Occupation: Software Engineer | Planet: Earth | Species: Magic Skeleton | Temperament: Twelve-tone equal | 🖥️🎵🧬🏳️🌈
Internet Explorer 🧭 Maker of linkitylink.lol
VIB Center for AI & Computational Biology
🧑💻 Director @steinaerts.bsky.social
📣 We're recruiting group leaders vib.ai
📻 Follow us for all the latest updates
America’s Finest News Source. A @globaltetrahedron.bsky.social subsidiary.
Get the paper delivered to your door: membership.theonion.com
Biosequence analysis using profile hidden Markov Models.
HMMER is managed by EMBL’s European Bioinformatics Institute (EMBL-EBI).
UCSD, HHMI
Neuroscience, protein design, climate change
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Postdoc Fellow @ Uni Neuchâtel 🇨🇭 | Transposable Elements, Annotation w/ Earl Grey, TE Evo Dynamics, Rapid Adaptation | #FirstGen | 🇬🇧🇫🇷 | Ski Touring | Biking | Join the TEsky community: https://bsky.app/profile/did:plc:mw4y54k2y45j
MLing biomolecules en route to structural systems biology. Asst Prof of Systems Biology @Columbia. Prev. @Harvard SysBio; @Stanford Genetics, Stats.
Fighting for budget priorities that reflect the values of families across the country.
Ranking Member @congressmanboyle.bsky.social
🌎 https://democrats-budget.house.gov

![\toprule
& & \multicolumn{4}{c}{CPU time / h} & & \multicolumn{4}{c}{Max RSS\tnote{a}{ } / GB} \\
\cmidrule(lr){3-6} \cmidrule(lr){8-11}
Read length / bp & & 24 & 100 & 200 & 24--200 & & 24 & 100 & 200 & 24--200\\
\midrule
Newmap & & 4 & 9 & 16 & 150.5 & & 7.6 & 7.6 & 7.6 & 7.6 \\
Genmap & & 2 & 1 & 1 & 275 & & 13.1 & 13.1 & 13.1 & 13.1 \\
Umap & & 31 & 42 & 57 & >4752\tnote{b} & & 7.8 & 7.5 & 5.9 & ${\sim}7.8$\tnote{c}\\
\bottomrule
\end{tabular}
\begin{tablenotes}
\item[a] Maximum resident set size.
\item[b] Estimate from a linear extrapolation of \qty{24}{\basepair} CPU time.
\item[c] Estimate from the largest maximum RSS found across all Umap benchmarks.
\end{tablenotes}](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:y6iikj3lebwl7k5cyi2fbahv/bafkreieliq6x7p4h6dqvxsgvzlitbptse3gl55qayic6qn3wqacsakxsd4@jpeg)