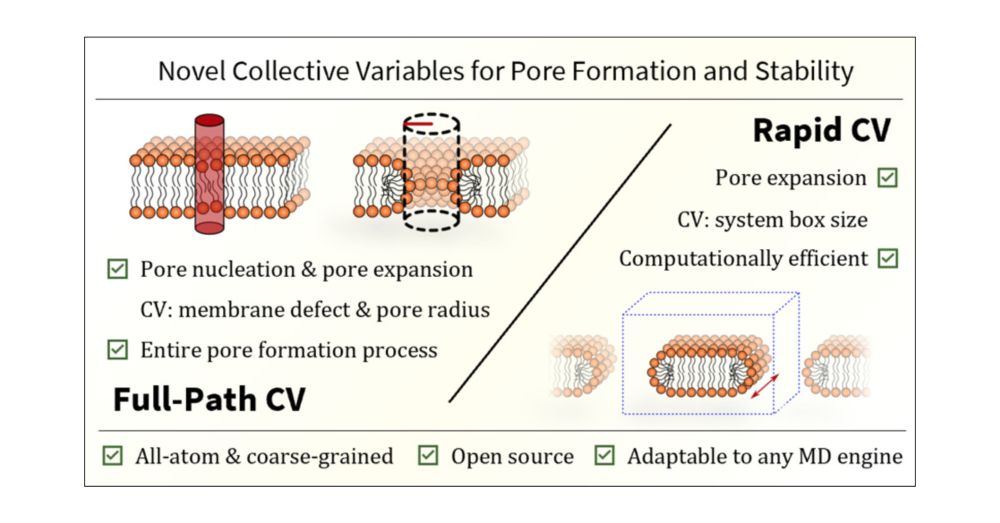
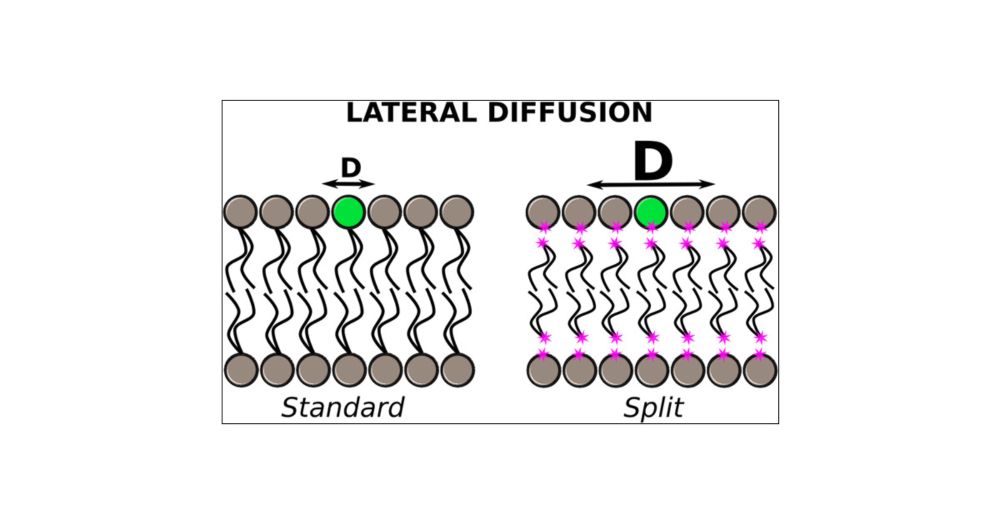
Redirecting
Second, we demonstrated how helical peptides can serve as sensors of membrane saturation: doi.org/10.1016/j.bp... Great work by Sushmita and Peter in collaboration with @javanainenm.bsky.social
07.10.2025 19:13 — 👍 1 🔁 0 💬 0 📌 0
Two new publications just dropped!
07.10.2025 19:13 — 👍 0 🔁 0 💬 1 📌 0
Now peer-reviewed and published! Dive in and enjoy. doi.org/10.1016/j.so...
17.07.2025 07:06 — 👍 0 🔁 0 💬 0 📌 0


Congrats to two new Ph.D. graduates: @ladme.bsky.social and Peter Pajtinka! Great defenses, great work, and great results all around. Big thanks to @javanainenm.bsky.social, Markus Miettinen, and Mario Vazdar for serving as opponents and visiting us in Brno.
13.07.2025 18:49 — 👍 0 🔁 0 💬 0 📌 0
Honored to receive this award!
26.05.2025 19:16 — 👍 3 🔁 0 💬 0 📌 0

Huge congrats to Sofia on a successful Ph.D. defense! 🎓👏 Wishing you all the best in your future endeavors. It's been a pleasure having you with us!
02.04.2025 09:20 — 👍 0 🔁 0 💬 0 📌 0
Follow the official CEITEC page for the latest news and cool science. 😎
11.03.2025 12:07 — 👍 1 🔁 0 💬 0 📌 0
A tahle story má pokračování, @mzenisek.bsky.social. 😊 Robert Vácha z @labvacha.bsky.social na CEITEC MUNI právě získal na navazující výzkum peptidů, ničitelů bakterií a rakovinných buněk, ERC Proof of Concept Grant! #sooooproud #supportscience!
23.01.2025 11:49 — 👍 2 🔁 1 💬 0 📌 0
We’re off to a strong start in 2025. First, work of Láďa on enhanced diffusion received a beautiful cover.
14.01.2025 16:55 — 👍 4 🔁 1 💬 1 📌 0