
African penguin
Source: Wildlife Conservation Society

Cotton top tamarin
Source: Wildlife Conservation Society

Eld's deer
Source: Wildlife Conservation Society

Elongated tortoise
Source: Wich’yanan (Jay) Limparungpatthanakij, via inaturalist.org and Wikimedia Commons
Thanks to the support of @wcs.org and Google Research, we have sequenced and assembled the genomes of nine endangered species, with more on the way!
To learn more: blog.google/innovation-a...
03.02.2026 15:50 — 👍 5 🔁 2 💬 0 📌 0

How we’re helping preserve the genetic information of endangered species with AI
Scientists are working to sequence the genome of every known species on Earth.
This blog talks about the great work of the
@ebpgenome.bsky.social. To support it Google.org has funded sequencing and open release of 13 genomes, with a $3M commit to sequence 150 more and develop methods to improve assembly finishing and other bottlenecks.
blog.google/innovation-a...
03.02.2026 01:50 — 👍 10 🔁 1 💬 0 📌 0

The killing of Alex Pretti is a heartbreaking tragedy. It should also be a wake-up call to every American, regardless of party, that many of our core values as a nation are increasingly under assault.
25.01.2026 17:39 — 👍 60234 🔁 19578 💬 3168 📌 1553

Hoiho - the world’s rarest penguin, fewer than 150 mainland pairs left
🐧We researched one of the world’s rarest #penguins. The yellow‑eyed penguin (aka hoiho/takaraka) isn’t one homogeneous species after all!
www.biorxiv.org/content/10.1...
#hoiho #conservation #genomics #birds #nzwildlife #endangered #wildlife #nature
28.10.2025 20:08 — 👍 59 🔁 21 💬 1 📌 3


Accurate somatic small variant discovery for multiple sequencing technologies with DeepSomatic www.nature.com/articles/s41... (read free: rdcu.be/eLny0) github.com/google/deeps...
16.10.2025 19:29 — 👍 11 🔁 5 💬 0 📌 1
Germline Small Variant Calling Workflow for SBX Duplex Data
Wednesday, September 10, 2025 at 12:00 PM Eastern Daylight Time.
I'll be speaking in this webinar (go.roche.com/sbx-d) on September 10, where I'll share our benchmarks and observations for Roche's SBX sequencing instrument, as well as models developed by our team for SBX data.
21.08.2025 00:18 — 👍 10 🔁 4 💬 1 📌 1
Also thanks to 20% contributors: Ben Soudry, Mike Kruskal, Sowmiya Nagarajan, Suchismita Tripathy, Francisco Unda, Vasiliy Strelnikov
And community contributions from Sam Yadav and Seraj Ahmad at Roche improving the code for custom model training
13.05.2025 20:23 — 👍 1 🔁 0 💬 0 📌 0

Release DeepSomatic 1.9.0 · google/deepsomatic
DeepSomatic:
In this release, we are introducing FFPE_WGS_TUMOR_ONLY and FFPE_WES_TUMOR_ONLY models.
The WGS and WGS_TUMOR_ONLY models have been retrained with all datasets described in the manusc...
Release led by Kishwar Shafin, contributions by Daniel Cook, Alexey Kolesnikov, Lucas Brambrink, and Pi-Chuan Chang as engineering manager.
Thanks to student researcher contributions from Farica Zhuang and Mobin Asri.
DeepSomatic release page:
github.com/google/deeps...
13.05.2025 20:23 — 👍 2 🔁 0 💬 1 📌 0

Release DeepVariant 1.9.0 · google/deepvariant
DeepVariant:
In this version we have updated our training scheme for the HG002 sample with the newly released HG002-T2T truth set which improves accuracy against that truth set.
Our labeling metho...
Release of DeepVariant and DeepSomatic v1.9
DV: Now train on HG002 T2T-Q100. Error reduction of 12% for Illumina and 30% for PacBio on this truth set. 25% faster. DeepTrio is 5x faster (20h -> 4h).
DS: New models FFPE_TUMOR_ONLY for {WGS, WES}. Much improved WGS models.
github.com/google/deepv...
13.05.2025 20:23 — 👍 20 🔁 9 💬 1 📌 0

Incredibly moving Justin Trudeau remarks:
"We have fought and died alongside you....During your darkest hours...we were always there. Standing with you, grieving with you, the American people....Canadians are a little perplexed as to why our closest friends and neighbors are choosing to target us."
02.02.2025 03:39 — 👍 22694 🔁 6244 💬 687 📌 445
You have some additional control on memory use by the number of threads you run with.
For running on GPU, I am not sure if you've seen this - github.com/google/deepv...
Which requires a little more configuration, but can let you better manage CPU-GPU tradeoffs. Definitely expert use.
01.02.2025 02:10 — 👍 0 🔁 0 💬 0 📌 0
Hi Eric, sorry to not notice till now. From the DV FAQ, we see the Keras model takes 16GB of memory (github.com/google/deepv...).
It's possible that pangenome-aware models will take more memory, and we do observe more memory per thread used for that. Definitely not lower than 16GB.
01.02.2025 02:06 — 👍 0 🔁 0 💬 2 📌 0
Great question. We were talking recently about L40S benchmarks. We don't have that data immediately on hand, but are planning to generate runtime stats for it.
06.12.2024 22:34 — 👍 1 🔁 0 💬 1 📌 0
They're very close - to the point that small changes of coverage or the inclusion of PCR in preparation would tip between one and the other.
06.12.2024 22:33 — 👍 3 🔁 0 💬 0 📌 0
Release led by DeepVariant tech lead Kishwar Shafin. Team Engineering manager Pi-Chuan Chang. Small model work led by Lucas Brambrink. Pangenome-aware led by Mobin Asri and Juan Carlos Mier. Fast pipeline by Alexey Kolesnikov. Kinnex/MAS-Seq model by Daniel Cook and Shiyi Yin from Verily. 3/3
05.12.2024 17:57 — 👍 2 🔁 0 💬 0 📌 0
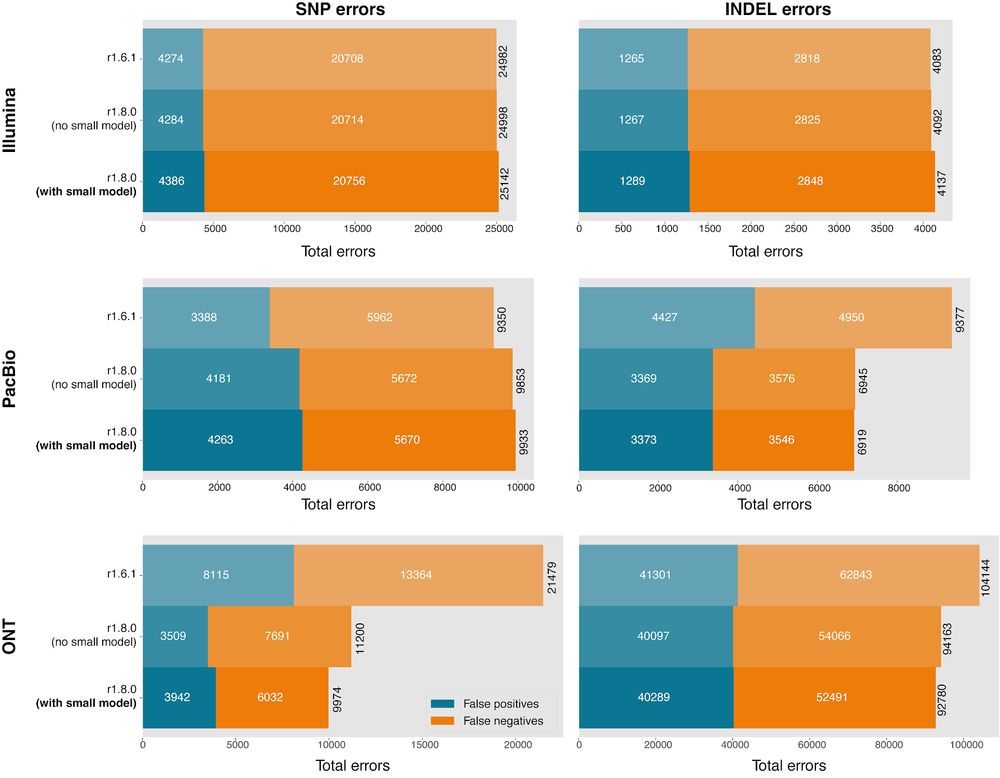
Plots of SNP and Indel error numbers for DeepVariant models. Shows a Indel error reduction of 26% for PacBio and a ~50% SNP error reduction for ONT.
Added SPRQ to PacBio training, reducing Indel error on SPRQ by 26%. Added Platinum Pedigree training data for PacBio model, reducing errors by 34% on more extensive Platinum truth. New model and case study for Kinnex/Mas-Seq/Iso-Seq. Additional speed options for GPU pipelines 2/3
05.12.2024 17:57 — 👍 8 🔁 4 💬 3 📌 0
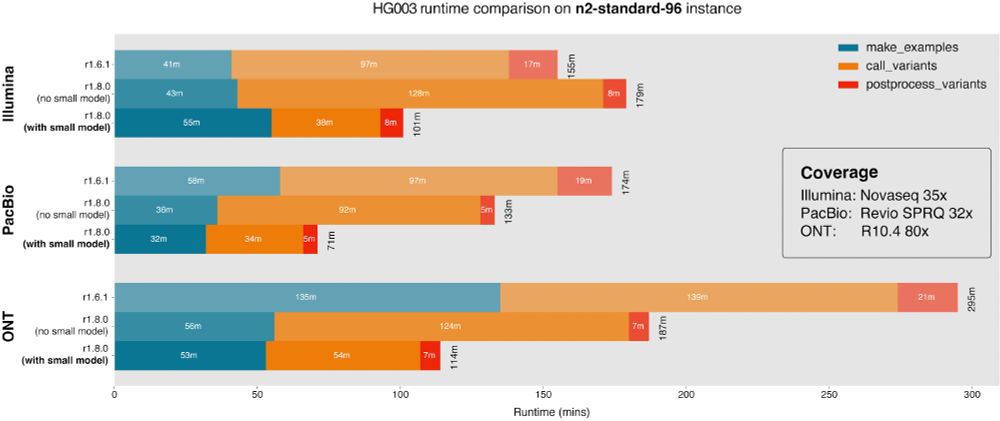
Runtime figure for new version of DeepVariant with and without small model. Showing reduction in runtime of 155 minutes to 101 minutes with Illumina, 174 minutes to 71 minutes with PacBio, and 295 minutes to 114 minutes with Oxford Nanopore.
Release of DeepVariant 1.8. Large speed improvement (~67% faster) via small model for easy sites. New Pangenome-aware option. Reduces error by ~30% for vg-mapped WGS, ~10% for BWA WGS, ~5% BWA exome. New config for custom model users, see release notes.
(github.com/google/deepv...)
05.12.2024 17:57 — 👍 38 🔁 14 💬 1 📌 0
Release by Kishwar Shafin
Major contributions from Pi-Chuan Chang, Daniel Cook, Alexey Kolesnikov
Google 20%ers: Will Kwan, Pauline Sho, Lucas Brambrink, Mo Samman, Atilla Kiraly
UCSC for vg: @benedictpaten.bsky.social, Shloka Negi, Jimin Park, Mobin Asri
Pacbio: Billy Rowell, Nathaniel Echols
26.10.2023 16:37 — 👍 1 🔁 0 💬 0 📌 0
There are now custom models and case studies for CompleteGenomics instruments.
T7: github.com/google/deepv...
G400: github.com/google/deepv...
For now, these are stand alone models. We'll likely consider whether we can jointly include these in the broad WGS model later.
26.10.2023 16:35 — 👍 2 🔁 0 💬 1 📌 0
The changes to DeepTrio for de novo detection are substantial. We now in two steps - first for overall accuracy and then a weighted fine tuning for de novos. Our benchmarks show large improvements in de novo calling relative to the prior DeepTrio.
github.com/google/deepv...
26.10.2023 16:34 — 👍 1 🔁 0 💬 1 📌 0
Want to benefit from pangenomes and want a recipe?
github.com/google/deepv...
Shows a step by step process, with Docker images for how to map to a Pangenome reference w/ vg and calls w/ DeepVariant. Final calls are more accurate and in GRCh38 coordinates. Thanks to the UCSC team for co-development
26.10.2023 16:34 — 👍 2 🔁 0 💬 1 📌 0

Release DeepVariant 1.6.0 · google/deepvariant
Improved support for haploid regions, chrX and chY. Users can specify haploid regions with a flag. Updated case studies show usage and metrics.
Added pangenome workflow (FASTQ-to-VCF mapping with V...
Release of DeepVariant v1.6.
Support for haploid regions, chrX/Y.
Workflow for Pangenome FASTQ-to-VCF.
Major DeepTrio improvements for de novo variants.
Models for CompleteGenomics T7, G400
Add NovaSeqX to training data
Release by Kishwar Shafin
github.com/google/deepv...
26.10.2023 16:32 — 👍 7 🔁 4 💬 1 📌 0
Proud of Ayse Keskus and Asher Bryant in my group for making this happen! This work is a collaboration with Children's Mercy, UCSC and Google Health - who are also releasing the first version of DeepSomatic today: github.com/google/deeps...
24.10.2023 16:57 — 👍 4 🔁 2 💬 1 📌 0

GitHub - KolmogorovLab/Severus: A tool for somatic structural variant calling using long reads
A tool for somatic structural variant calling using long reads - GitHub - KolmogorovLab/Severus: A tool for somatic structural variant calling using long reads
TThanks to Jimin Park and Benedict Paten from UCSC, Mikhail Kolmogorov from NCI for analysis and testing. This group has also developed Severus for somatic SV calling, and we've worked closely with them
(github.com/KolmogorovLa...)
Thanks to Khi Pin Chua and Billy Rowell from PacBio
24.10.2023 16:54 — 👍 2 🔁 0 💬 0 📌 0

GitHub - google/deepsomatic
Contribute to google/deepsomatic development by creating an account on GitHub.
Initial release of DeepSomatic, which identifies subclonal variants when given tumor and normal BAM files. Pre-trained models and case studies available for Illumina and PacBio. Development led by Kishwar Shafin which built off a framework by Pi-Chuan Chang.
(github.com/google/deeps...)
24.10.2023 16:53 — 👍 36 🔁 17 💬 2 📌 1
Best resource for getting extracellular domain localization from Ensembl gene/protein IDs?
I tried the subcellular locations in HPA but the membrane annotation is mostly fully intracellular proteins.
🧪🧬🖥️🔬
14.09.2023 01:41 — 👍 8 🔁 4 💬 3 📌 0
Evolutionary biologist, computational biologist, statistician. I like to develop mathematical models of evolutionary process and see how they fit to data. I also like cities where building apartments is legal.
Oncologist-scientist at Weill Cornell and New York genome center. Into somatic evolution, trees and words
Geneticist, researcher and educator
Genome annotation geek at University of Greifswald, always interested in friendly scientific collaboration. Developer of BRAKER, GALBA, MakeHub and other Gaius-Augustus tools.
Gemma Product Manager @google DeepMind
- Gemma 💎
- Google AI ⚙️🧠
The Vertebrate Genomes Project (VGP) aims to generate error-free genome assemblies for all 70,000 vertebrate species, offering a powerful resource to advance research in biology, genomics, conservation, medicine, and bioinformatics.
Colorado RNA biologist & tRNA enthusiast exploring the wild frontiers of nanopore direct RNA sequencing at the intrepid venn diagram of northern blots & machine learning.
Passionate scientists leveraging genomics to understand diseases and seeking clinical applications
Computational biologist 🧬🖥️ at Mass General Brigham and Broad Institute, PhD at Harvard. Bioinformatics, transcriptomics, COVID, genomics, immunology, genetics, statistics, and web development. I made #ggrepel and I blog at https://slowkow.com
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
🇨🇦 boiling in the Arizona desert and not riding my bikes enough. Cancer researcher focused on multiple myeloma and clinical genomics at TGen, part of City of Hope. Creator of MyelomaSky and Sequencing Tech feeds. Director, Collaborative Sequencing Center
Space cowboy riding a massive rock across the galaxy, wrangling genomes and herding data. PhD in Biochemistry @TAMUBCBP. Working @LanzaTech.
GitHub: https://github.com/dbrowneup
LinkedIn: https://linkedin.com/in/drbrowne
Behavioral geneticist, dad, cyclist, reluctant dog owner, formerly young, allegedly irreverent, unlike most people my age, I was raised in the Great Depression.
Nucleic acid explorer and psychedelic music lover. I still Believe In Peace 🕊️Chemistry Prof@TelAvivUni, Founder of JaxBio
http://www.nanobiophotonix.com
https://jaxbio.com/
Schmidt AI in Science Postdoctoral Fellow at U. Chicago
theoretical & computational soft matter, biophysics, machine learning
jordanshivers.github.io
Computational Neuroscientist, PhD.
https://katiaplopes.github.io/


















