Thanks for putting this together - some really great reads!
22.12.2025 12:50 — 👍 1 🔁 0 💬 0 📌 0
🚨 Want to do a #Phd using high-throughput mutagenesis and CRISPR screens to understand how gene body methylation impacts transcription in collaboration with Greg Kudla's group? Do apply, deadline in a week! www.findaphd.com/phds/project...
Deadline 15th December 2025
#epigenetics Please share 🙏
08.12.2025 09:33 — 👍 7 🔁 7 💬 1 📌 1

Boundary issues: SWI/SNF shapes chromatin patterns in and around centromeres
Abstract. The SWI/SNF family of chromatin remodelling complexes, comprising BAF, PBAF and ncBAF, is known for their critical roles in regulating chromatin
Some Friday reading...new review in Open Biology where we discuss how SWI/SNF maintains the chromatin environment around centromeres, building on recent work from ourselves and others. Lots of fun putting this together with @alison-harrod.bsky.social and @thedownslab.bsky.social!
05.12.2025 10:29 — 👍 13 🔁 5 💬 0 📌 1
Thank you Takashi!!
30.11.2025 12:35 — 👍 1 🔁 0 💬 0 📌 0

Heterochromatin plays a key role in restricting the position of the CENP-A/C domain, ensuring only one CENP-A is nucleated per centromere. SUV39H1/2 and SETDB1 H3K9me3 methyltransferases differentially control neocentromeres, pericentromeric, and core alpha satellites. 4/4
27.11.2025 21:58 — 👍 2 🔁 0 💬 0 📌 0

We discover that CENP-A chromatin at a de novo human neocentromere and at canonical alpha satellites are highly plastic! And are surrounded by dense regions of H3K9me3 heterochromatin (& DNAme)! But is this functional? 3/4
27.11.2025 21:58 — 👍 1 🔁 0 💬 1 📌 0
With new developments in single molecule epigenomics using @nanoporetech.com, we revisited a key question in chromosome biology - why does pericentric heterochromatin surround the centromere? We hypothesised that it may be required to keep an epigenetic centromere locus in place! 2/4
27.11.2025 21:58 — 👍 2 🔁 0 💬 1 📌 0

differential contribution of H3K9 methyltransferases to boundaries at satellites
A new and fascinating story from @bencarty.bsky.social and the group, with crucial help from the teams of @naltemose.bsky.social, Simona Giunta, and @dfachinetti.bsky.social. Many thanks to all for a fantastic collaboration.
www.nature.com/articles/s41...
25.11.2025 12:16 — 👍 66 🔁 32 💬 10 📌 1
Congrats Dani, Nick and team!!
04.09.2025 18:39 — 👍 1 🔁 0 💬 0 📌 0
A fantastic review of the cell cycle control of centromeres from our lab. Lots learned and many open questions 40 years later!
29.07.2025 19:42 — 👍 1 🔁 0 💬 0 📌 0
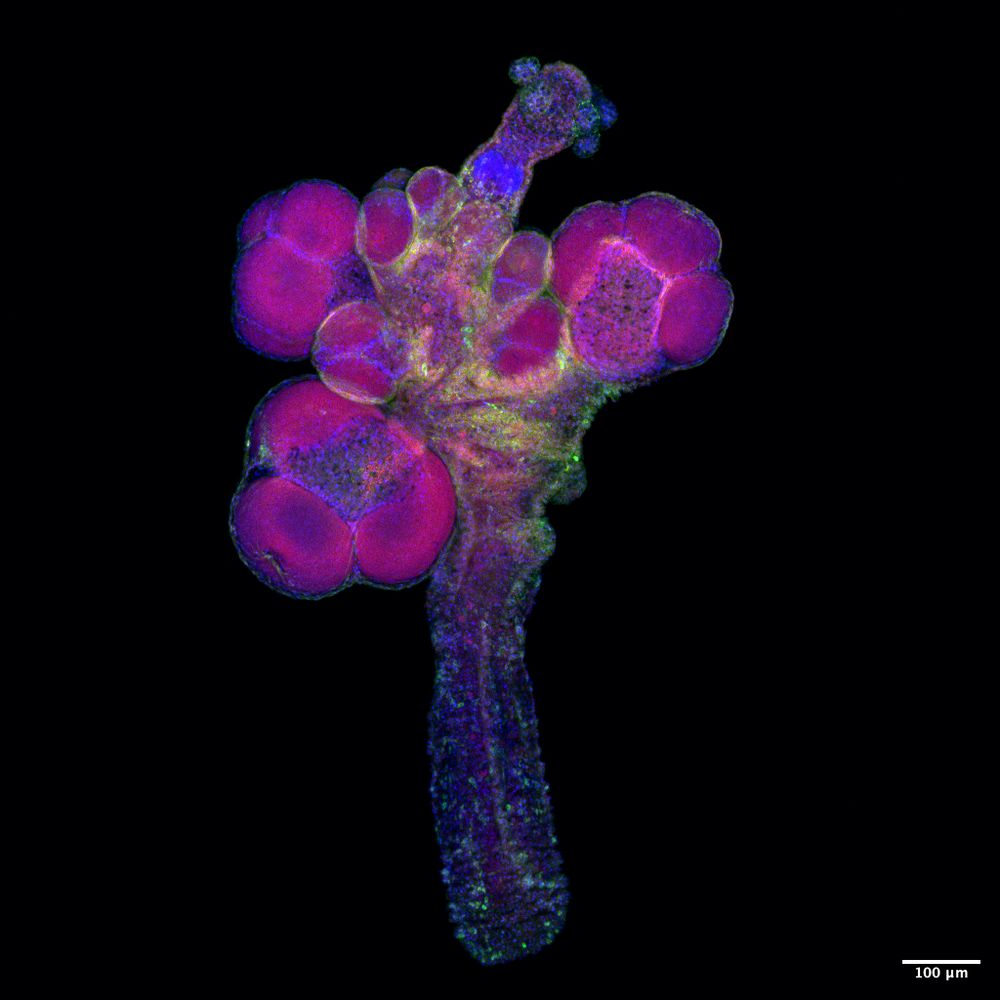
I am looking for a new postdoc to join the lab. Interested in pluripotency, germ cells, and in investigating these in a genetically tractable cnidarian? Get in touch! We offer a long-term contract, excellent research environment, and a lovely city #Galway. www.urifranklab.org
20.06.2025 13:24 — 👍 54 🔁 36 💬 1 📌 2
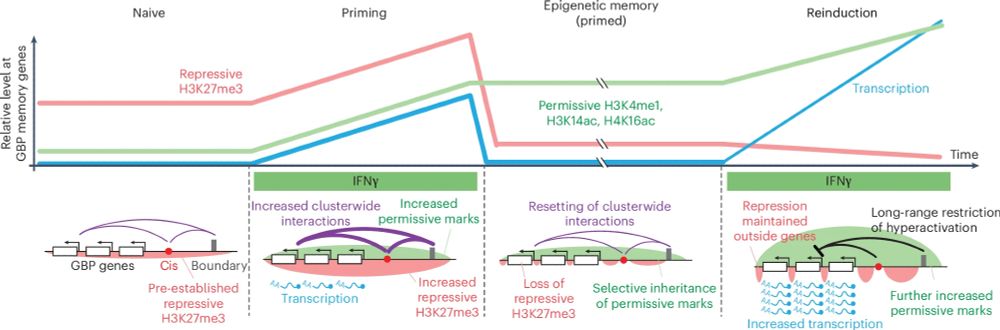
Fresh of the press! rdcu.be/egmY0 We discovered key chromatin modifications that are heritably maintained through mitotic cell divisions in the absence of continued transcription. These hold a memory of past exposure to interferon-gamma, priming cells for future activation of gene expression.
04.04.2025 10:22 — 👍 10 🔁 6 💬 1 📌 0

Unlocking the Pause: A new player in gene transcription
A new work from the Klose lab published in the journal Molecular Cell
Research @oxfordbiochemistry.bsky.social @ox.ac.uk supported by the Wellcome Trust (UK) and NIH (USA) reveals essential new player in the process of deciding whether genes should be made or not. Great work @jesskelley.bsky.social
@robklose.bsky.social @edimitrova.bsky.social 👏 bit.ly/202502-klose
10.02.2025 13:50 — 👍 17 🔁 9 💬 0 📌 0

This was a very collegial and great collaboration between @jansenlab.bsky.social @naltemose.bsky.social fachinetti and Giunta Labs, which exemplifies what's great about our centromere community. Stay tuned!
08.02.2025 19:10 — 👍 2 🔁 0 💬 0 📌 0

But what about the rest of the alpha satellites? To our surprise, we nucleate new centromeres on satellite arrays when we reduce H3K9me3 repressive chromatin. New CDRs, and what appear to be functional dicentrics on mitotic chromosomes!
08.02.2025 19:10 — 👍 1 🔁 0 💬 1 📌 0

But does this matter? Absolutely YES. Centromeres move and expand, sometimes quite dramatically.
08.02.2025 19:10 — 👍 0 🔁 0 💬 1 📌 0

But how is H3K9me3 being regulated at centromeres? We find roles for SETDB1, SUV39 H1/H2, as well as non-canonical roles for PRC2 component SUZ12! Disruption has a substantial effect on H3K9me3 boundaries
08.02.2025 19:10 — 👍 0 🔁 0 💬 1 📌 0

So what about canonical centromeres? First, we find that the active centromere locus contains a distinct dip in H3K9me3, analogous to 5mCG Centromere Dip Regions!
08.02.2025 19:10 — 👍 1 🔁 0 💬 1 📌 0

Neo4p13 formed in a SUV39 regulated H3K9me3 domain. We perturbed H3K9me3 and (compensating) H3K27me3 surrounding this centromere.. with some quite dramatic effects on the neocentromere!
08.02.2025 19:10 — 👍 1 🔁 0 💬 1 📌 0

How can we model centromere position and inheritence? Using our neocentromere in RPE1 cells (Neo4p13).. we assess centromere position over 100 days of culture. It moves.. somewhat.. but overall size is remarkably consistent!
08.02.2025 19:10 — 👍 0 🔁 0 💬 1 📌 0
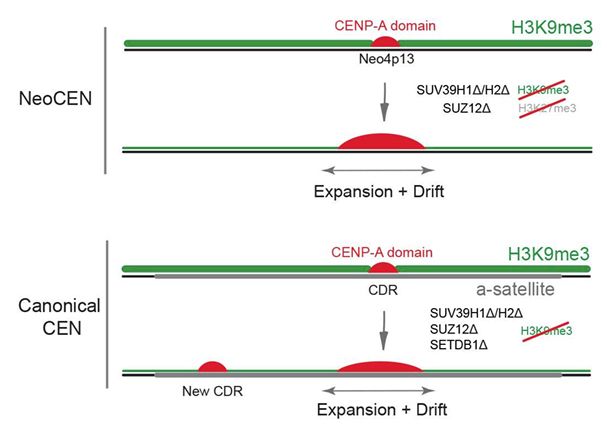
@bencarty.bsky.social and the team did a fantastic job in a wonderful collaboration with @naltemose.bsky.social as well as Dani Fachinetti and Simona Giunta’s teams to help define the molecular basis for maintaining centromeric chromatin size and position. www.biorxiv.org/content/10.1...
05.02.2025 17:33 — 👍 38 🔁 14 💬 4 📌 2
Professor for Genetics interested in chromatin structure, histone variants and histone modifications and their roles in gene regulation and disease development.
Senior editor @naturesmb.bsky.social. Views and opinions are personal.
Postdoc @icr.ac.uk.bsky.social in @thedownslab.bsky.social
🔬🧪🥼👩🏻🔬🧫
Postdoc @Rugg-Gunn lab @Babraham Institute & Newnham College Cambridge | Prev. Wellcome trust DPhil student @BrockdorffLab @UniofOxford | Passionate about mammalian developmental epigenetics, sc-genomics, CRISPR screens, science outreach and communication.
Prof of Computational Microbiology. Oxford Biochemistry & St Anne’s College. Bacterial Cell Envelopes. LFC fan 🌈
The Downs lab at the Institute of Cancer Research in London, studying chromatin biology and genome stability. thedownslab.org
Interested in all things chromosome segregation and aneuploidy | Director & PI at Hubrecht Institute
Group leader of transposon group in Oxford at IDRM. #new_pi https://sites.google.com/view/berrenslab
Epigenetics & Chromatin Biology | Polycombs in Development & Disease | Cancer Genetics | Professor of Chromatin Biology, Trinity College Dublin | www.brackenlab.com
Postdoc @Gijs Wuite lab @vuamsterdam.bsky.social
Previously EMBO Postdoctoral fellow @nynkedekkerlab.bsky.social @ox.ac.uk
PhD @Michael Nash lab @unibas.ch
Single-molecule biophysics | DNA replication | Protein/DNA/chromosome mechanics
I run, ski, and engineer cells and materials; asking how nutrient sensing & signaling work: bit.ly/RK-GoogleScholar
🇪🇪: TalTech| Ex: Tartu Ülikool; 🇸🇪: Karolinska Institutet, Chalmers; 🇯🇵: Kyutech; 🇮🇳: IITD
Assistant Professor at University of Texas at San Antonio. Study evolution/regulation of selfish genes. Manage two labs. Research lab🔬🧪🧬🖥️ with some ease but the other lab 🐶🐾 is un-manageable!
https://www.anti-sense.org/
Chromatin biochemistry and genomics in development and cancer. UW-Madison School of Medicine and Public Health
TheLewisLab.net
Teif lab at the University of Essex. We work on gene regulation in chromatin and applications to liquid biopsies, using approaches of genomics, biophysics, bioinformatics & AI. Our focus is nucleosomics, TF binding, CTCF, cfDNA. https://generegulation.org
We study chromosomal replication & recombinational repair mechanisms, Smc5/6, SUMO-based controls using multidisciplinary approaches. Tweets are a group effort.
Postdoctoral researcher in Mia Levine’s Lab, University of Pennsylvania
Love all things Drosophila, evolution, running and Eastenders
Oncologist @DukeU; interested in cancer research, clinical trials, immunotherapy but many other things - Views are my own
Thekhasrawlab.com



















