
Out now: we’re happy to share our new preprint on CroCoNet (Cross-species Comparison of Networks), a framework for robust comparative network analyses.
👉 doi.org/10.1101/2025...
@boyanbonev.bsky.social
Group Leader at Helmholtz Center Munich * Brain Epigenomics

Out now: we’re happy to share our new preprint on CroCoNet (Cross-species Comparison of Networks), a framework for robust comparative network analyses.
👉 doi.org/10.1101/2025...

Our new manuscript, led by Emily Corrigan, examines inhibitory neuron diversity across approximately 160 million years of evolutionary divergence, as part of BRAIN Initiative Cell Atlas Network (BICAN) developing brain atlas package: www.nature.com/articles/s41...
07.11.2025 18:06 — 👍 60 🔁 22 💬 2 📌 1🚀 𝗣𝗼𝘀𝘁𝗱𝗼𝗰 𝗢𝗽𝗲𝗻𝗶𝗻𝗴
We’re recruiting an ERC-funded Postdoctoral Researcher to explore how 3D gene regulatory networks evolved during human and primate brain development, using cutting-edge single-cell multiomics, organoid models, and CRISPR screens.
👉 Apply by Nov 30 | www.bonevlab.com/join
A wonderful collaboration with @alexbaffet.bsky.social @boyanbonev.bsky.social and @neurogenesis-i3s.bsky.social labs
28.10.2025 07:18 — 👍 4 🔁 2 💬 0 📌 0
Our latest research is online in the @biorxivpreprint.bsky.social! 🧬🎉
🔎 We have developed a novel, single-cell real-time approach to monitor R-loop generated TRCs, and we have shown R-loop-mediated, replisome-controlled transcription repression.
www.biorxiv.org/cgi/content/...

🎤 Apply to speak @our TALENT FORUM by July 31!
Showcase YOUR research in Epigenetics & NucleicAcids @www.helmholtz-munich.de & connect with leading scientists
🧳Travel+accommodation covered if selected!
📝 Info+how to apply: lnkd.in/drkRvz7N
#postdoc #career #groupleader #HelmholtzMunich #research
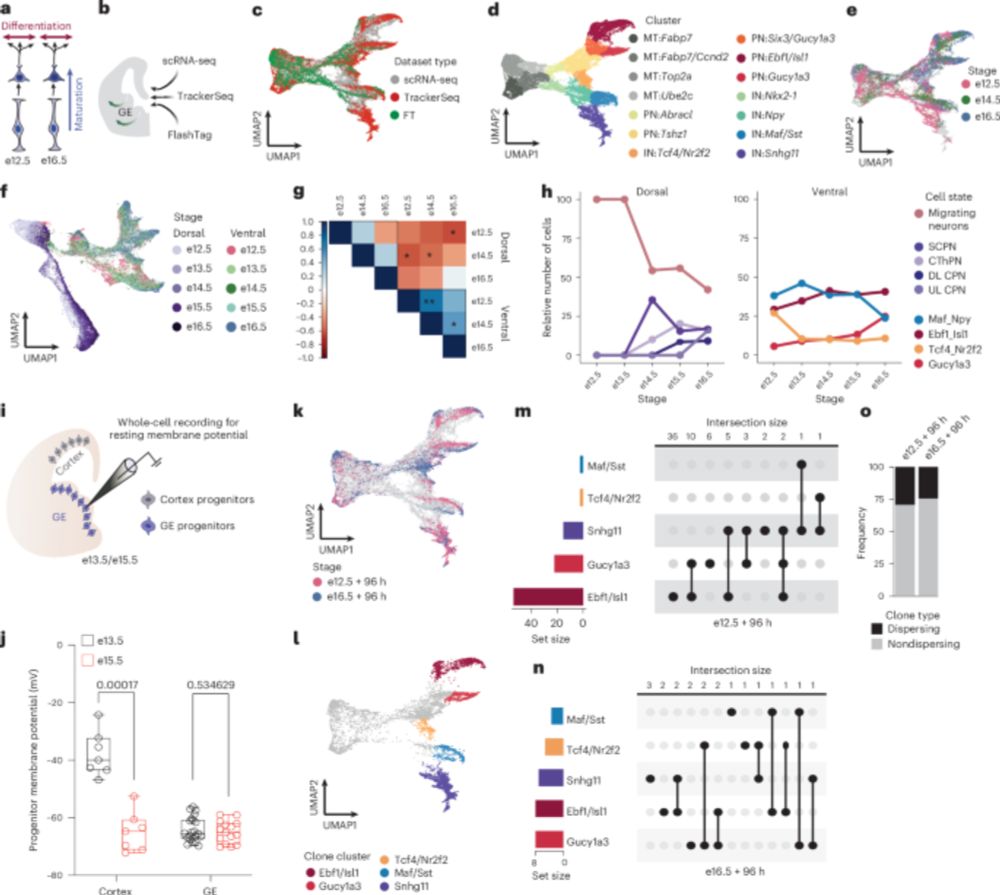
Now out in @natneuro.nature.com: Temporal control of progenitor competence shapes maturation in GABAergic neuron development in mice. Congrats to first authors @annrosebright.bsky.social, Yana Kotlyarenko, @flo-neuhaus.bsky.social, and thanks to all collaborators!
www.nature.com/articles/s41...
1/n
Are you @ the late stages of your postdoc ? Want to pursue a PI career ?
Then this 👇👇 is for you ! Apply for a spot @ our Talent Forum
www.helmholtz-munich.de/en/stem-cell...
Peer-networking / Career orientation and more !!!
Please distribute :) 🙂
Looking forward to it!
15.05.2025 04:59 — 👍 1 🔁 0 💬 0 📌 0
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
Check out our latest work on the evolution of animal genome regulation out today in @nature.com. Nicely summarized below by @ianakim.bsky.social.
www.nature.com/articles/s41...
This is a major output from our ERC-StG project Evocellmap @erc.europa.eu at @crg.eu

In the most recent episode we talked with @boyanbonev.bsky.social from @bonevlab.bsky.social at @epihmgu.bsky.social about his work on neuroepigenetics, focusing on gene regulation, chromatin architecture, and primate epigenome evolution. #epigenetics
Listen here: activemotif.com/podcasts-boy...
This was a great collaboration between the @bonevlab.bsky.social and the lab of Amos Tanay, led by Yonatan and Florian with substantial contribution from @silviavangelisti.bsky.social, Faye and Aviezer. If you found this study of interest, please RT, comment and share! All feedback is welcome. 14/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 0 📌 0Code and data to reproduce all figures, or browse and reanalyze, is available at github.com/tanaylab/mmcortex, either separately or in a standalone docker container. You can also explore the data interactively at apps.tanaylab.com/MCV/mmcortex/. 13/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Finally, we assayed the activity of thousands of CREs isolated from their genomic context across multiple timepoints in vivo using a cell-type-specific massively parallel reporter assay. We showed that cell type-specificity is encoded in the sequence, in agreement with our quantitative model. 12/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
We also show explicitly that NSC methylation has a repressive effect on IPC accessibility, and that cooperativity between proximal CREs is correlated with accessibility both in terms of sequence content and total proximal activation. 11/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0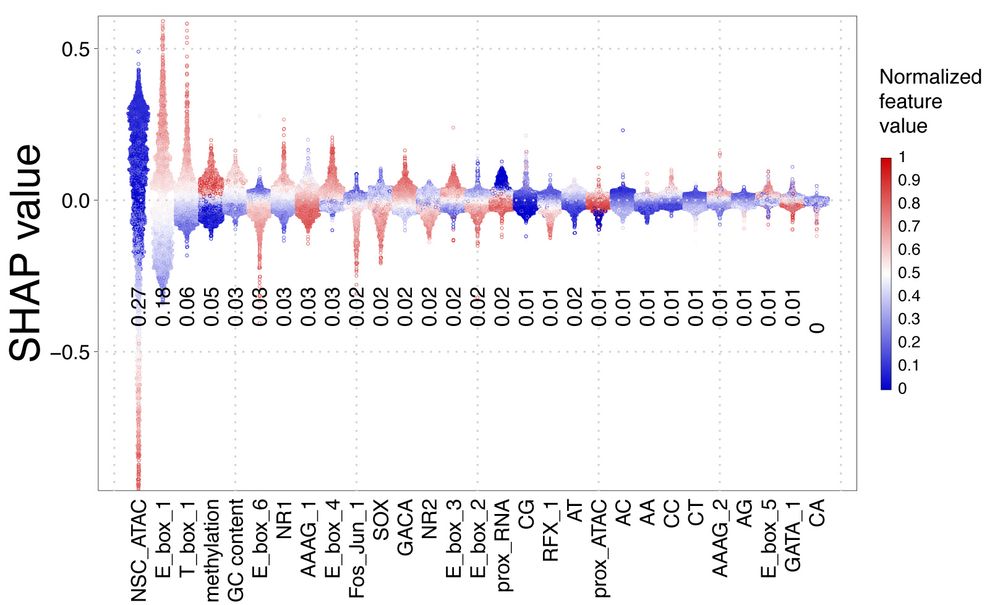
We developed a machine-learning model that integrates sequence (putative TF motifs, dinucleotide content) and epigenomic features (ATAC, methylation, proximal activity) to predict NSC<->IPC specificity of CREs, that yields R^2 = 0.62 on held-out data. 10/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Moreover, temporally activating CREs in NSCs get compacted significantly closer over time to astrocyte- or NSC-specific TSSs, an effect not observed with IPC TSSs. 9/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Our Hi-C analysis shows that while overall chromatin topology is conserved across time in NSCs, there are specific hotspots whose differential insulation across time is correlated with the presence or depletion of proximal CREs. 8/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Temporally activating CREs are significant: they are enriched with astrocyte-specific CREs, and astro CREs undergo extensive demethylation within NSCs across time, in contrast to other cell type-specific CREs. 7/14
04.04.2025 16:37 — 👍 2 🔁 0 💬 1 📌 0
We found that ATAC-activating CREs are tightly concomitant with methylation decrease. But the opposite was not true! Deactivating CREs in NSCs stay unmethylated across our time series, and this is not due to absence of DNMTs. 6/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Furthermore, we were able to identify CREs with dynamic accessibility within cell types, across time. We focused on CREs that either activate/increase accessibility, or deactivate over time in NSCs. 5/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Our metacell model of ATAC states provides a high-resolution timeline of cis-regulatory element (CRE) accessibility in corticogenesis. We use it to show that CREs specific to corticofugal neurons are activated late in maturation, in stark contrast to callosal projection neuron-specific CREs. 4/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
We show that astrocyte genes in NSCs are upregulated gradually over time, not suddenly, along with erosion of NSC proliferative capacity. This erosion is also correlated with lengthening of the cell cycle, suggesting that NSC fate bias is coupled to the cell cycle. 3/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
We have generated one of the most comprehensive multiome datasets in the context of brain development, integrating time-series measurements of scRNA-seq, scATAC-seq, 5mC methylation and Hi-C of mouse somatosensory cortex across 6 developmental timepoints. 2/14
04.04.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Can a stem cell change its epigenome while proliferating? In collaboration with the lab of Amos Tanay, we demonstrate that yes, the epigenome of neural stem cells (NSCs) is continuously remodeled, across multiple layers, during mouse corticogenesis. 1/14
www.biorxiv.org/content/10.1...
I'm happy to share our work on the neurodevelopmental impact of ADNP mutations in ASD and beyond. It has been an exciting ride to share with @alessandrovitriolo.bsky.social, Mariana, and the people of the Testa and Poot Labs.
Full paper here:
www.biorxiv.org/content/10.1...
Thread below 🧵
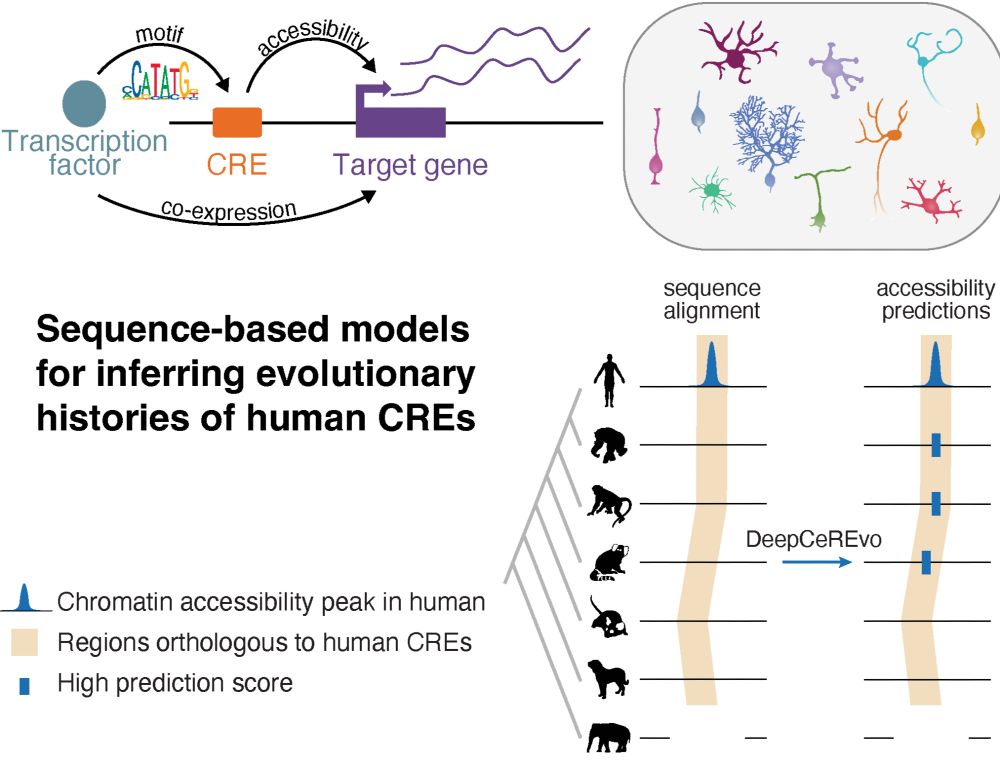
How does gene regulation shape brain evolution? Our new preprint dives into this question in the context of mammalian cerebellum development! rb.gy/dbcxjz
Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social
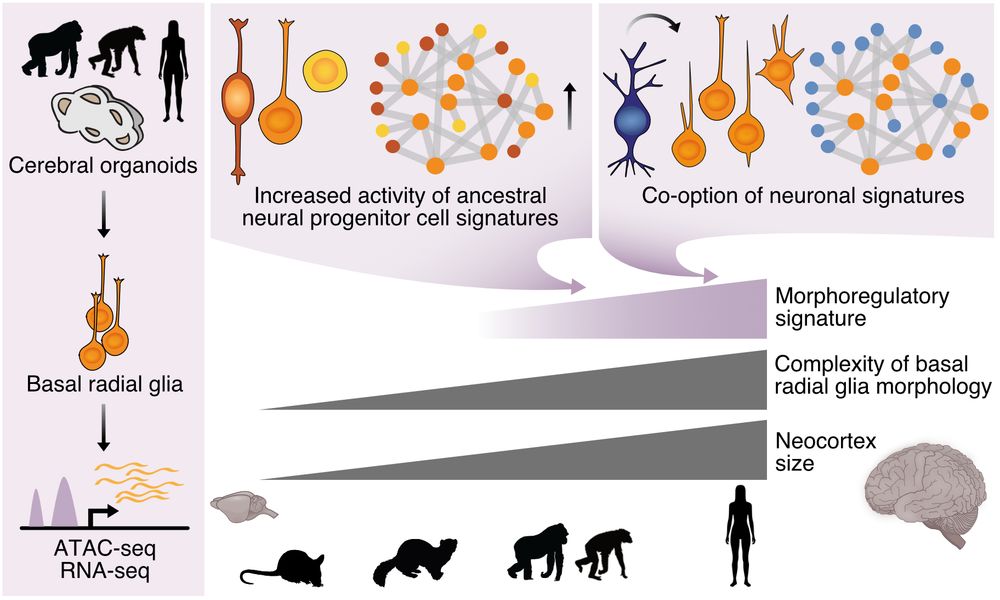
💫 New preprint from the lab: Comparing great ape cerebral organoids, we found that human-specific morphoregulatory signatures in basal radial glia characterise neocortex evolution. Fantastic work from super-talented PhD student Theresa Schütze!
www.biorxiv.org/content/10.1...
This was a heroic effort by my very first PhD student - Silvia, with a significant contribution from other lab members and a great collaboration with the Enard lab @enardhellmannwg.bsky.social. Stay tuned for more exciting research from the @bonevlab.bsky.social very soon!
13.03.2025 09:59 — 👍 3 🔁 0 💬 1 📌 0