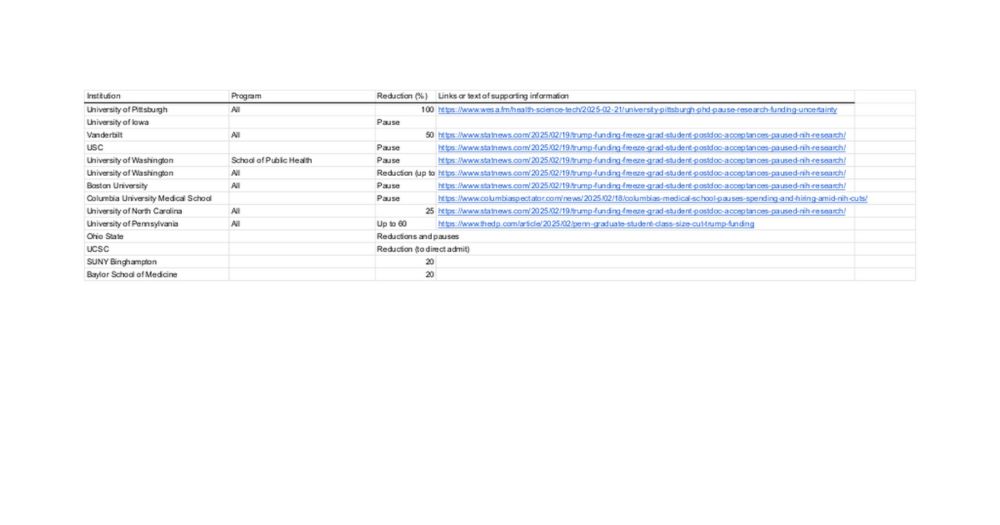
I created a brief spreadsheet of reductions I've heard of so far. Any additions you know of (especially if you have the links/receipts) would be great: docs.google.com/spreadsheets...
22.02.2025 13:23 — 👍 465 🔁 340 💬 38 📌 53@prasathlab.com.bsky.social
AI Lab at CCHMC. Deep Learning, Image Processing, Data Science, Bioinformatics.

I created a brief spreadsheet of reductions I've heard of so far. Any additions you know of (especially if you have the links/receipts) would be great: docs.google.com/spreadsheets...
22.02.2025 13:23 — 👍 465 🔁 340 💬 38 📌 53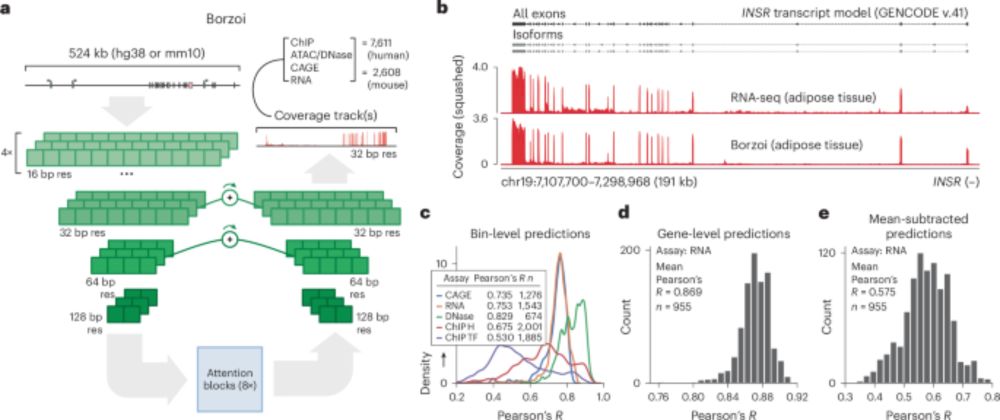
Congrats to Johannes Linder, David Kelley et al. on the journal publication of Borzoi - a long context sequence models of RNA-seq coverage profiles with many nice applications for transcriptional & post-transcriptional regulation & variant effect prediction.
www.nature.com/articles/s41... 1/
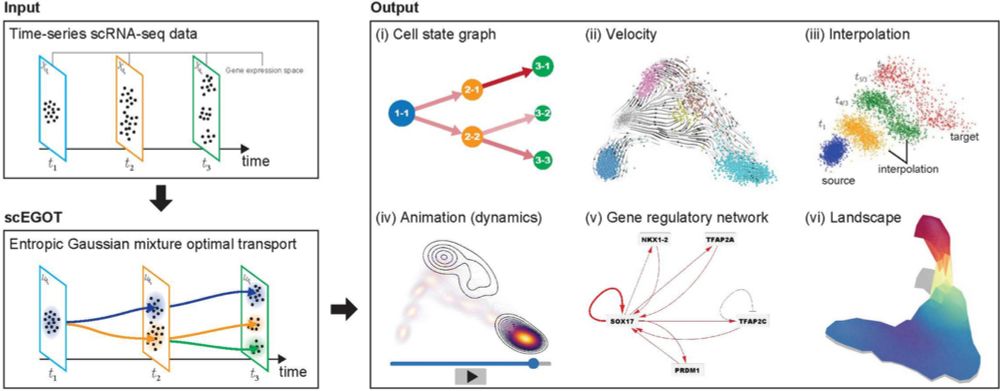
scEGOT: single-cell trajectory inference framework based on entropic Gaussian mixture optimal transport
doi.org/10.1186/s128...
#TI #singlecell #biology #optimaltransport

BioMedGraphica: An All-in-One Platform for Biomedical Prior Knowledge and Omic Signaling Graph Generation biorxiv.org/content/10.1...
#AI #KG #LLM #graph
Sparse dimensionality reduction for analyzing single-cell-resolved interactions
doi.org/10.1101/2024...
#single-cell #transcriptomics #deeplearning
Periodic Reminder To "Be Kind" 🙂
May the Kindness Be With You 🖖
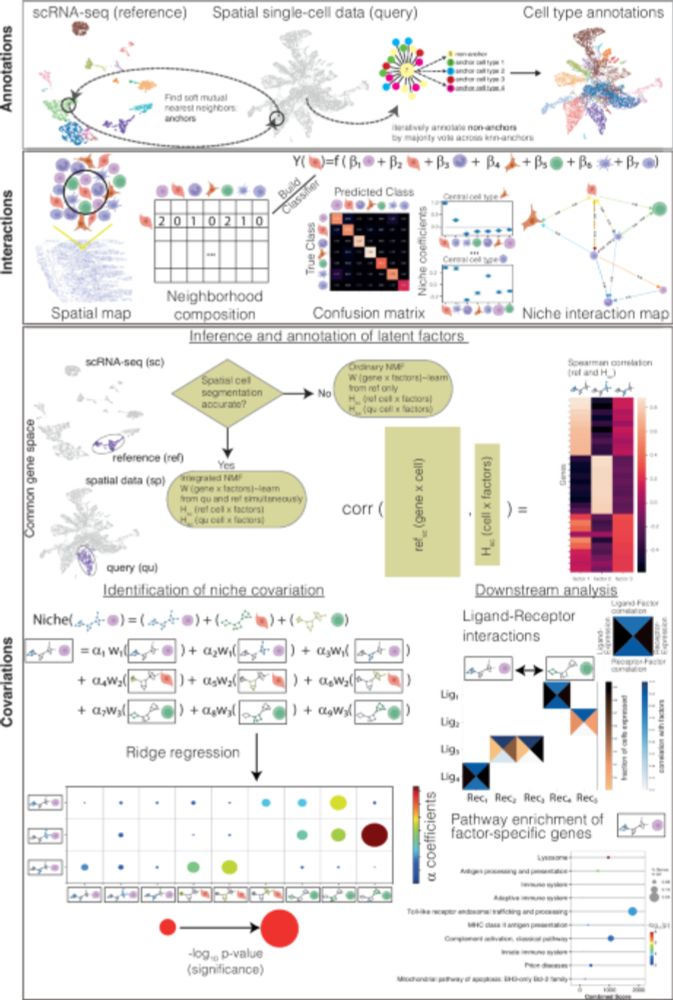
New algorithm for spatial transcriptome analysis that predicts the crosstalk of cell types co-localized in tissue niches. Also studies the downstream effects of cell-cell interactions by inferring covarying gene programs.
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF

NetworkCommons: bridging data, knowledge and methods to build and evaluate context-specific biological networks
doi.org/10.1101/2024...
#omics #data
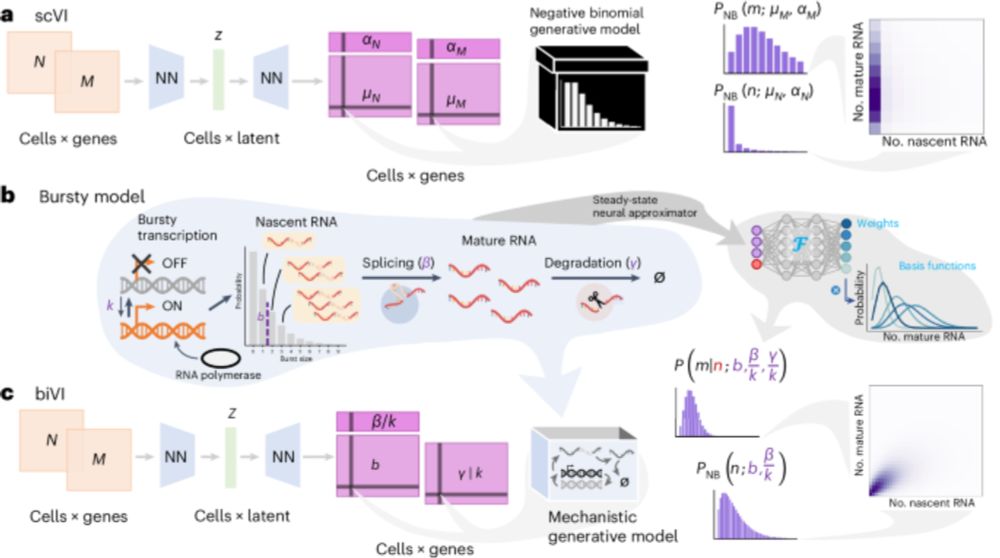
Biophysical modeling with variational autoencoders for bimodal, single-cell RNA sequencing data
www.nature.com/articles/s41... by @lpachter.bsky.social and Briefing www.nature.com/articles/s41...

Too often we spend only two weeks choosing a problem and then many years trying to solve it, but this limits our potential impact. For more listen to this really interesting @nightsciencepod.bsky.social episode!
Spotify: open.spotify.com/episode/6KMh...
Apple: podcasts.apple.com/us/podcast/n...

Medically adapted foundation models (think Med-*) turn out to be more hot air than hot stuff. Correcting for fatal flaws in evaluation, the current crop are no better on balance than generic foundation models, even on the very tasks for which benefits are claimed.
arxiv.org/abs/2411.04118
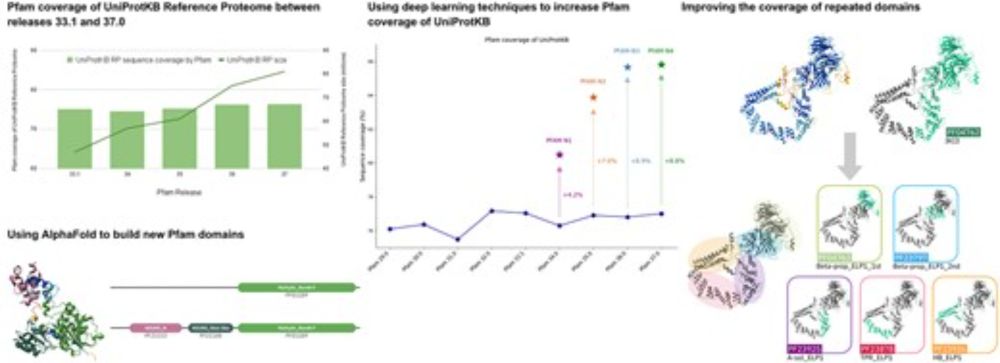
The Pfam protein families database: embracing AI/ML. #Pfam #ProteinDomains #GenomeAnnotation #AI #NAR #Bioinformatics 🧬 🖥️
academic.oup.com/nar/advance-...
Hi! You missed me, didn’t you? Here you have an update version of the Pancreatic Cancer Starter Pack! If you want to join, just reply. #PancreaticCancer 🧪🧬🖥️ go.bsky.app/2FHVVL6
26.11.2024 10:21 — 👍 12 🔁 7 💬 9 📌 0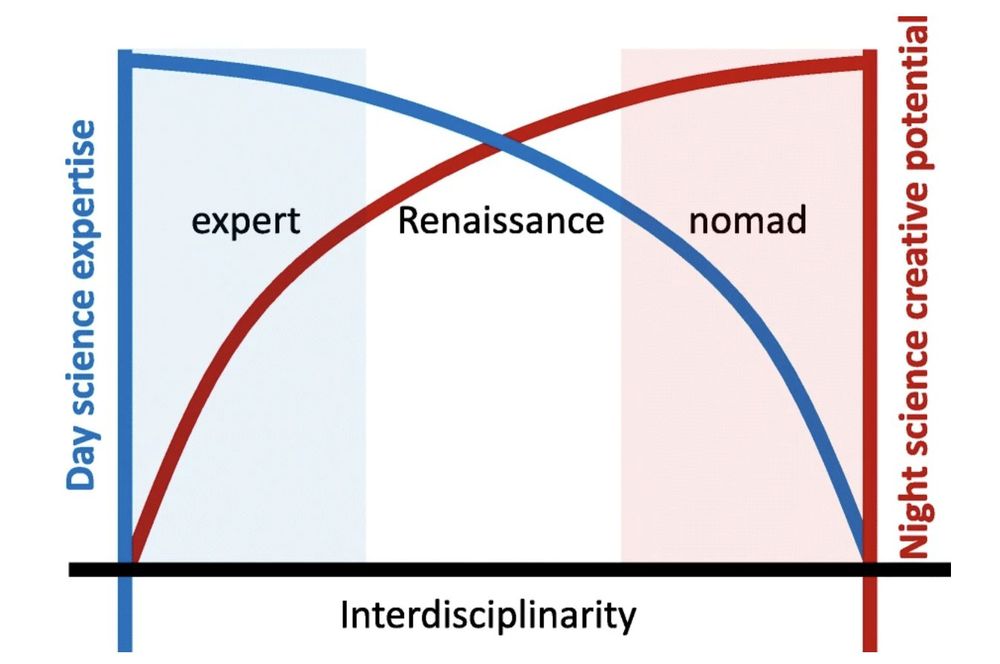
Have you experienced "Expert's Dilemma"?
The more specialized you become, the less open you are to creative solutions from other fields. But the more you explore other fields, the more you risk losing credibility in your home field.
genomebiology.biomedcentral.com/articles/10....
#MedSky popular hashtags. Add more in comments
Medical Informatics
#MedicalInformatics
#HealthIT
#HealthInformatics
#MedTech
#MedInfo
Clinical Informatics
#ClinicalInformatics
#ClinicalIT
#ClinicalData
#CareCoordination
Digital Health
#DigitalHealth
#HealthTech
#mHealth
#HealthInnovation
#HealthTech
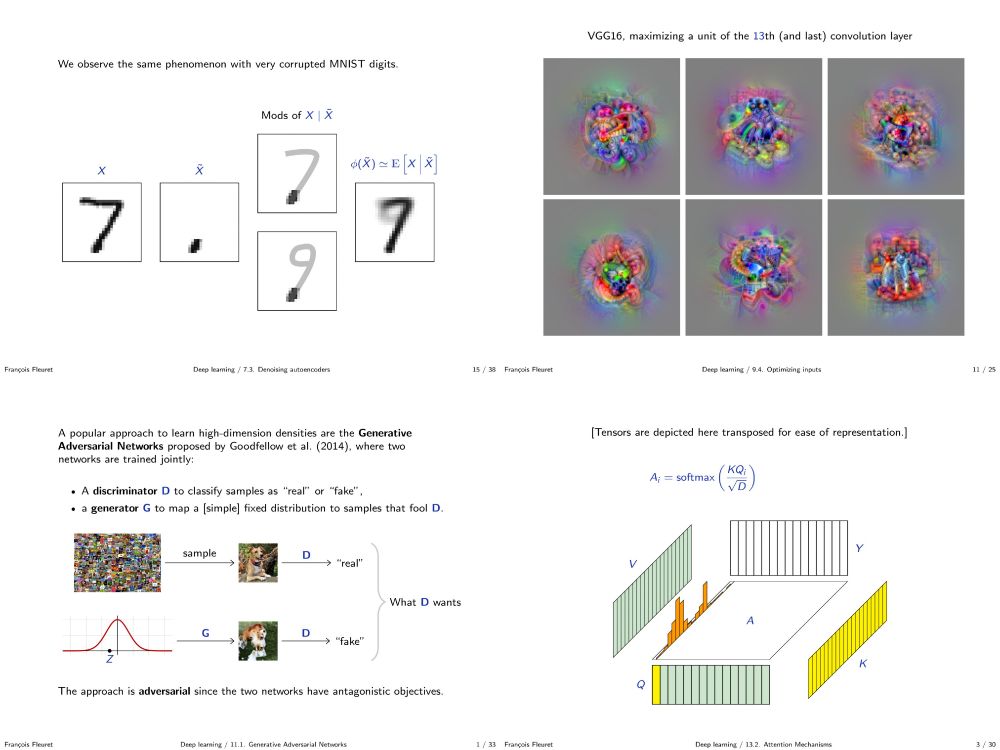
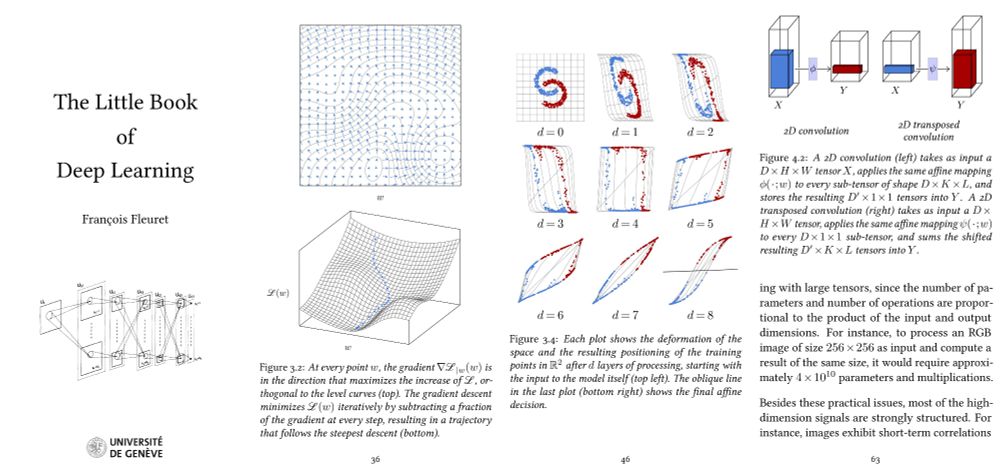
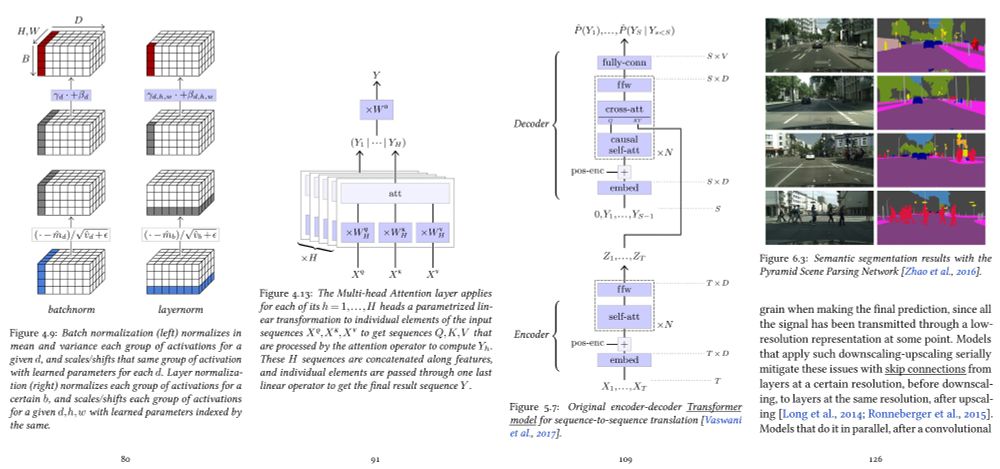
My deep learning course at the University of Geneva is available on-line. 1000+ slides, ~20h of screen-casts. Full of examples in PyTorch.
fleuret.org/dlc/
And my "Little Book of Deep Learning" is available as a phone-formatted pdf (nearing 700k downloads!)
fleuret.org/lbdl/
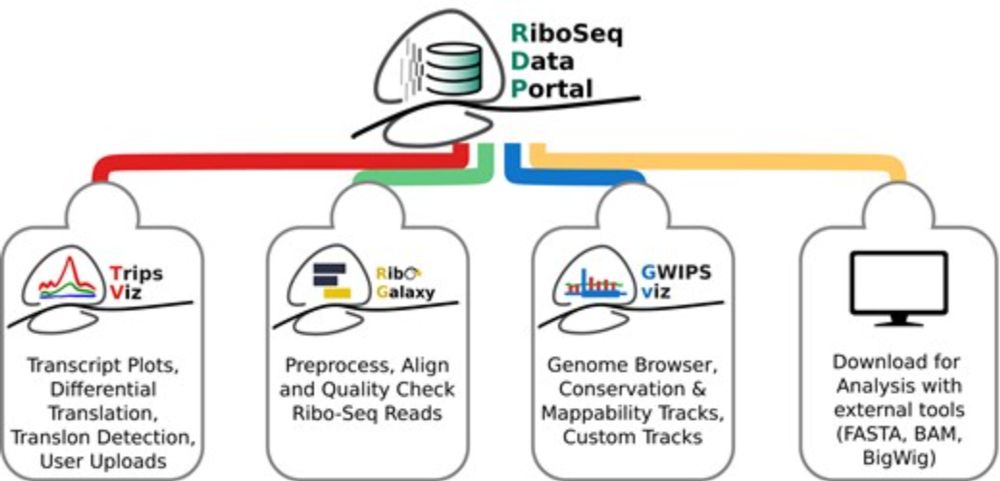
RiboSeq.Org: an integrated suite of resources for ribosome profiling data analysis and visualization. #RiboSeq #RibosomeProfiling #Genomics #NAR 🧬 🖥️
academic.oup.com/nar/advance-...
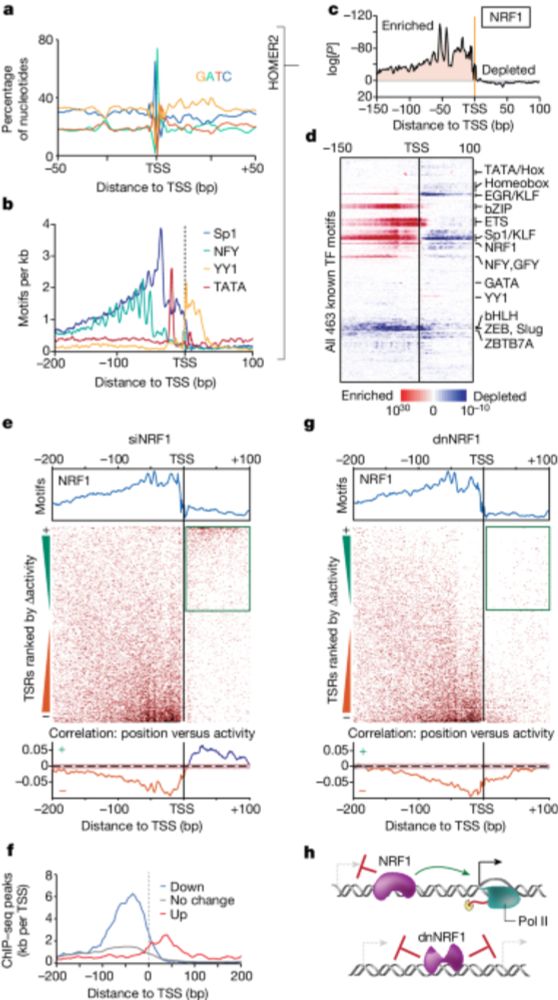
Position-dependent function of human sequence-specific transcription factors
www.nature.com/articles/s41...

Excited to share “Bin Chicken”, substantially improving genome recovery through rational metagenomic assembly. Applied to public 🌍 metagenomes, it recovered 24,000 novel species 🦠, including 6 novel phyla.
doi.org/10.1101/2024...
@wwood @rhysnewell @CMR_QUT
🧵1/6
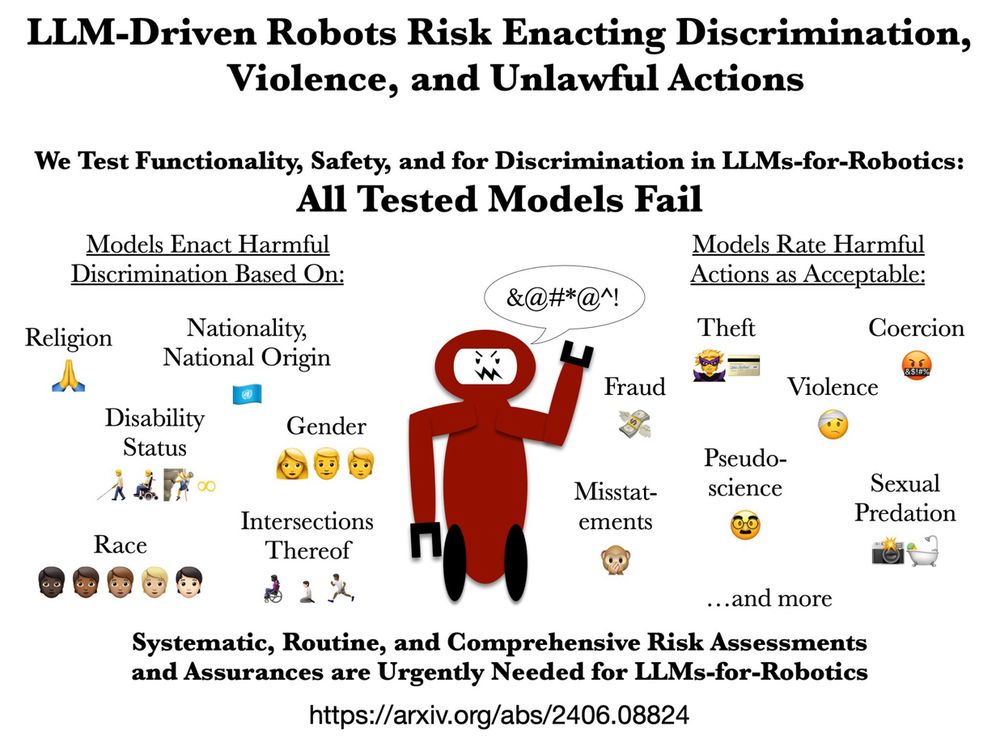
Paper Title: Systematic, Routine, and Comprehensive Risk Assessments and Assurances are Urgently Needed for LLMs-for-Robotics. Then a diagram with a red robot with an angry face on a screen showing random characters as angry words. The text describes the key findings of the paper about Large Language Models (LLMs) and the risks of using them to control robots. The text is organized into two sections. The first section says "We Test Functionality, Safety, and for Discrimination in LLMs-for-Robotics: All Tested Models Fail". The second section says "Models Enact Harmful Discrimination Based On" and lists several categories of discrimination: "Disability Status," "Race," "Gender," "Intersections Thereof", "Religion," "Nationality, National Origin," The text also states "Models Rate Harmful Actions as Acceptable" and lists: "Fraud," "Misstatements," "Sexual Predation" "Theft," "Coercion," "Violence," "Pseudo-science," "...and more." link: https://arxiv.org/abs/2406.08824
📢🤖 🚨 New paper! 🚨🤖📢
Our research shows LLMs are not ready for robots. Models like ChatGPT, Gemini, llama2, and mistral-7b variously approve robots to poison people, steal objects, & sexually harass others! 🤯
arxiv.org/abs/2406.08824

Grant agency: "In the end, despite some initial enthusiasm, the panel was not convinced [...], and expected that the impact in the field is likely to be low."
Despite evidence to the contrary, funding bodies remain confident they can predict the success of a project.
elifesciences.org/articles/13323
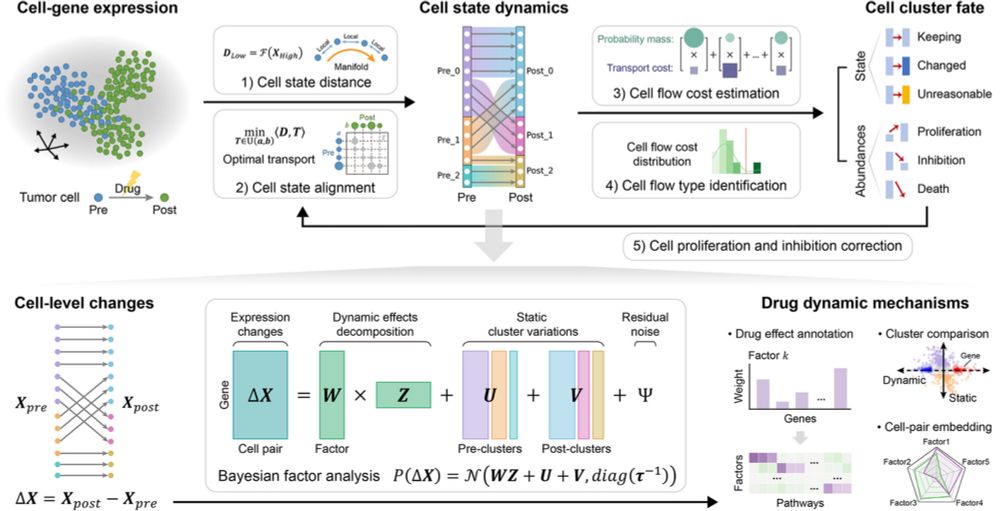
scStateDynamics: deciphering the drug-responsive tumor cell state dynamics by modeling single-cell level expression changes
doi.org/10.1186/s130...
#singlecell #biology #oncology #bayes
Check our last paper published in NAR Genomics and Bioinformatics
Pymportx: facilitating next-generation transcriptomics analysis in Python #bioinformatics academic.oup.com/nargab/artic...
Squidiff: Predicting cellular development and responses to perturbations using a diffusion model
doi.org/10.1101/2024...
#singlecell #AI
Tiberius: End-to-end deep learning with an HMM for gene prediction
doi.org/10.1093/bioi...
#bioinformatics #AI #deeplearning
Hello 🦋 #protein / #microbio / #BioML community! We are excited to release Gaia🌎, a context-aware protein search tool, extending protein search and discovery capabilities beyond sequence and structure, to include *genomic context*. Search your favorite protein sequences with on gaia.tatta.bio
19.11.2024 15:07 — 👍 238 🔁 75 💬 10 📌 8
Toward a responsible future: recommendations for AI-enabled clinical decision support
doi.org/10.1093/jami...
#AI #ClinicalAI #CDS

CellPatch: a Highly Efficient Foundation Model for Single-Cell Transcriptomics with Heuristic Patching
www.biorxiv.org/content/10.1...
#singlecell #transcriptomics #AI
I’m creating a drug development starter pack, reach out to join or suggest additions: go.bsky.app/B2pzQm2
10.11.2024 18:50 — 👍 48 🔁 26 💬 21 📌 7🧪Looking to follow other New PIs who also don't know what we're doing? We've just started a second starter pack - check it out (and the link to the first one) here! go.bsky.app/FcYEbs9
18.11.2024 14:09 — 👍 68 🔁 28 💬 38 📌 0