
Burrows-Wheeler Indexing - YouTube
Videos on : (a) the Burrows-Wheeler Transform (BWT), (b) the FM Index, which uses the BWT to construct a full-text index, (c) Wheeler graphs, (d) r-index, an...
I've added 7 videos to my Burrows-Wheeler indexing playlist (www.youtube.com/playlist?lis...), rounding out the r-index series and adding a 5-part series on the move structure. Now 27 videos in that playlist. I aim to add videos on prefix-free parsing, PBWT, Wheeler languages/automata in the future.
07.10.2025 14:17 — 👍 62 🔁 15 💬 2 📌 1
@acarroll.bsky.social @kishwar.bsky.social Pi-Chuan Chang, Faith Okamoto, @humanpangenome.bsky.social
02.10.2025 06:40 — 👍 1 🔁 0 💬 0 📌 0
Big thanks to everyone involved in the paper! @benedictpaten.bsky.social @adamnovak.graphs.vg Jordan Eizenga @jltsiren.bsky.social @jmonlong.bsky.social @shlokanegi.bsky.social Francesco
Andreace @sagorika03.bsky.social @kokyriakidis.bsky.social Glenn Hickey, Stephen Hwang, Emmanuele Delot
02.10.2025 06:40 — 👍 2 🔁 0 💬 1 📌 0

A cartoon of a variation graph and a sequencing read with seed alignments between the read and graph. The snarl tree corresponding to the variation graph is shown to the right. Below is the zip code tree representing the seeds, snarl and chain boundaries, and the distances among them.
My favorite part: co-linear chaining in cyclic graphs using zip code trees, a new data structure for quickly finding distances in the graph that best match corresponding distances in the read
02.10.2025 06:40 — 👍 6 🔁 0 💬 1 📌 0
Also featuring a cool demo of pangenome-guided regional assembly of complex loci from @shlokanegi.bsky.social Francesco Andreace @sagorika03.bsky.social @kokyriakidis.bsky.social
02.10.2025 06:31 — 👍 3 🔁 0 💬 1 📌 0
🦒Long read giraffe is out!🦒
Mapping long reads to pangenome graphs is ~10x faster than with GraphAligner, with veeery slightly better mapping accuracy, short variant calling, and SV genotyping than GraphAligner or Minimap2
02.10.2025 06:28 — 👍 42 🔁 22 💬 1 📌 0
Republicans are shutting down the government to stop you from having healthcare—there’s the message!!!!
01.10.2025 14:47 — 👍 1 🔁 1 💬 0 📌 0

Last talk of the day (before posters) "Lossless Pangenome Indexing Using Tag Arrays" presented by Parsa Eskandar! #WABI25
20.08.2025 19:59 — 👍 11 🔁 3 💬 0 📌 0
Thanks so much!
22.03.2025 07:07 — 👍 0 🔁 0 💬 0 📌 0
Huge thank you to my incredible advisor @benedictpaten.bsky.social at @ucscgenomics.bsky.social for all his support and guidance, and to my committee David Haussler, @russcd.bsky.social, and @jianma.bsky.social
21.03.2025 23:02 — 👍 4 🔁 0 💬 1 📌 0

I'm very excited to share that I defended my thesis this week! I helped develop algorithms to combat reference #bias and make genomics more #equitable and #representative by incorporating genetic #diversity into genomics pipelines
21.03.2025 23:00 — 👍 9 🔁 1 💬 1 📌 0
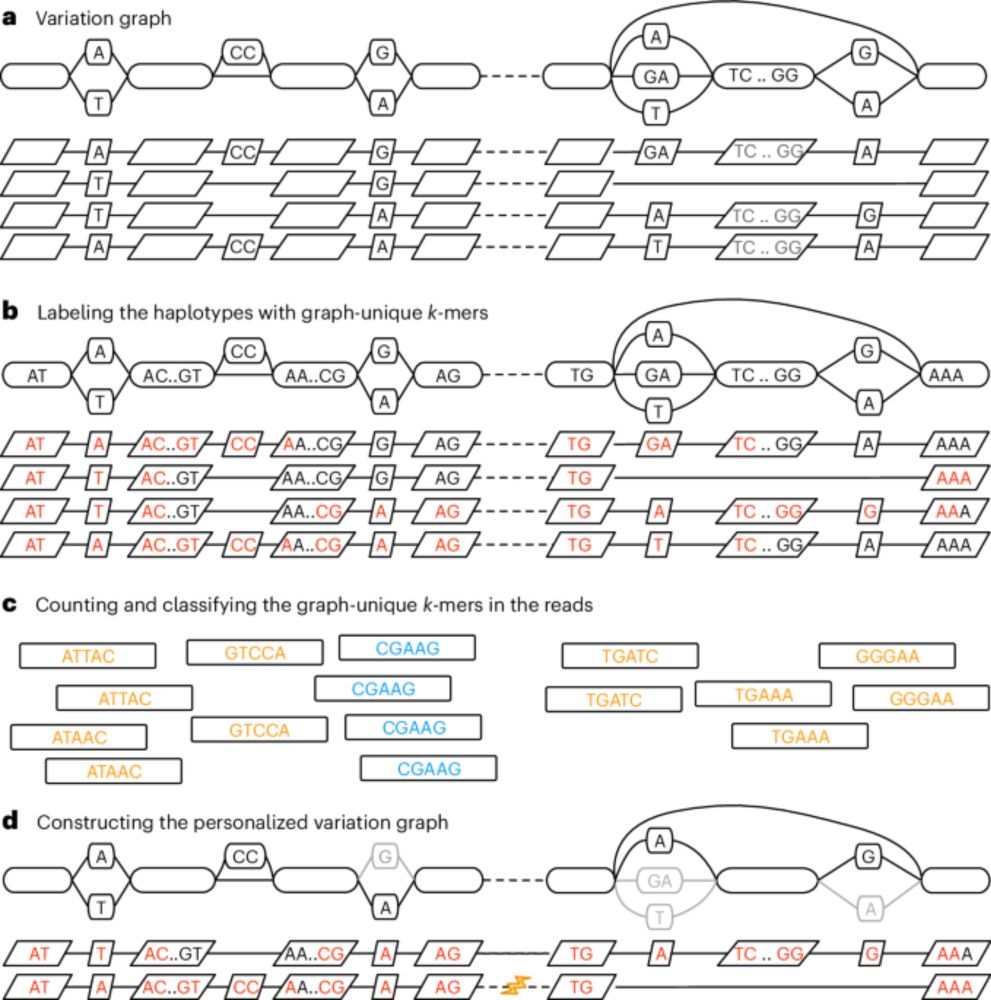
Personalized pangenome references - Nature Methods
This work introduces a k-mer-based approach to customizing a pangenome reference, making it more relevant to a new sample of interest. This method enhances the accuracy of genotyping small variants an...
We use personalized references with our Giraffe aligner. Each chromosome is partitioned into a sequence of blocks. We sample the most relevant haplotypes in each block using kmer counts. Mapping to this personalized reference improves variant calling accuracy. www.nature.com/articles/s41...
04.03.2025 23:00 — 👍 17 🔁 8 💬 1 📌 0

Welcome to the Bluesky account for Stand Up for Science 2025!
Keep an eye on this space for updates, event information, and ways to get involved. We can't wait to see everyone #standupforscience2025 on March 7th, both in DC and locations nationwide!
#scienceforall #sciencenotsilence
12.02.2025 17:04 — 👍 11519 🔁 5452 💬 291 📌 672
Variation graph representing the names of long read pangenome mappers: MiniGraph, GraphChainer, GraphAligner, PanAligner, and Giraffe
New figure for my thesis showing why Giraffe is the best long read pangenome mapper. No further evidence will be provided.
14.02.2025 19:45 — 👍 4 🔁 1 💬 1 📌 0

Quote: Long read sequencing is likely the next best test for unsolved cases... It can serve as a single diagnostic test, reducing the need for multiple clinical visits and transforming a years-long diagnostic journey into a matter of hours. Shloka Negi, UC Santa Cruz
Researchers at @ucsantacruz.bsky.social have demonstrated how long-read sequencing could improve detection of diseases that have eluded diagnosis, at a fraction of the cost. New clinical tests could be on the way.
🗞 news.ucsc.edu/2025/01/pate...
👏🏼👏🏿👏🏾 @khmiga.bsky.social @benedictpaten.bsky.social
24.01.2025 19:17 — 👍 15 🔁 7 💬 0 📌 1
Hi!
I'm Marion, a PhD student in the Barber lab at the University of Jena. I am interested in human pathogenic fungi, pangenomics, transcriptomics, and a sprinkle of evolution. Also fascinated about astrobiology ✨
I love large biomedical data.
Using genomic information to improve medicine
PhD fellow in immunogenetics @ University of Oslo |
I like ripped genes, long genes, baggy genes and double genes
Geneticist whose lab does experimental evolution, using yeast as a model. Because being a footballer was never going to work out, due to lack of talent.
PI of SGD and CGD
ORCID: 0000-0002-1692-4983
🌱 PhD student at Weigelworld @plantevolution.bksy.social | Graph pangenome | Population genetics
@downingtim@genomic.social - also downingtim.bsky.social - Head of #Genomics at The #PirbrightInstitute (UK). He/him. All posts (etc) are in a personal capacity. I log in regularly, nearly every month. #Andacyclist #genomics #virus #pathogen #evolution
Research group in Lille, Fr.
PhD student in algorithmic bioinformatics at @bonsaiseqbioinfo.bsky.social.
Interested in randomized algorithms and space-efficient data structures
https://igor.martayan.org
Associate Professor at the Department of Mathematics, Stockholm University, and a Scilifelab Fellow. Algorithms, Modeling, Transcriptomics, Genomics.
Hobby runner 5000m 18:48 | 10k 37:40 | HM 1:27:43 | M 3:39:06
Genome informatics at the Regeneron Genetics Center. All opinions, etc. my own. she/her 🇵🇭🏳️🌈
PhD student at @univlille.bsky.social
Graph visualisation for RNA and DNA sequences and databases
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
Professor, Molecular & Cellular Biology, Harvard University
Ph.D. candidate @WashUdbbs @WashUGenetics
Graduate student @ Computational Genomics Lab, UCSC Genomics institute
Graduate Student Researcher,
Computational Genomics Lab,
UC Santa Cruz
Associate Professor
DFCI & HMS








